Comparison of metabolic maps?
It is possible and it is automated!
Comparisons of target and source metabolic information most often suffers from the lack of experimental data. That is why metabolic simulators are often used.
On the other hand, due to complexity of the simulated metabolic maps, the similarity between them is difficult to be estimated by hand in side-to-side comparison.
Several modules, newly implemented in OASIS software, have been designed to automate different aspects of metabolic similarity that can be gleaned for comparisons of metabolic maps. Of particular importance are found to be the identification of common metabolic pathway(s), common metabolite(s), and similarity concerning the formation of reactive metabolites. Furthermore, the module could compare them taking into account their frequency of application, quantity of the producsed metabolites or just their appearance. The metabolic similarity results are presented in a transparent way.
The justified similarity in the metabolic maps of the target and source chemicals will aid in establishing metabolic consistency between the chemicals used in a read-across argument (i.e., establish similarity in their (a)biotic transformations) and it will strengthen the prediction.
- Metabolic similarity with respect to the simulated (a)biotic transformations
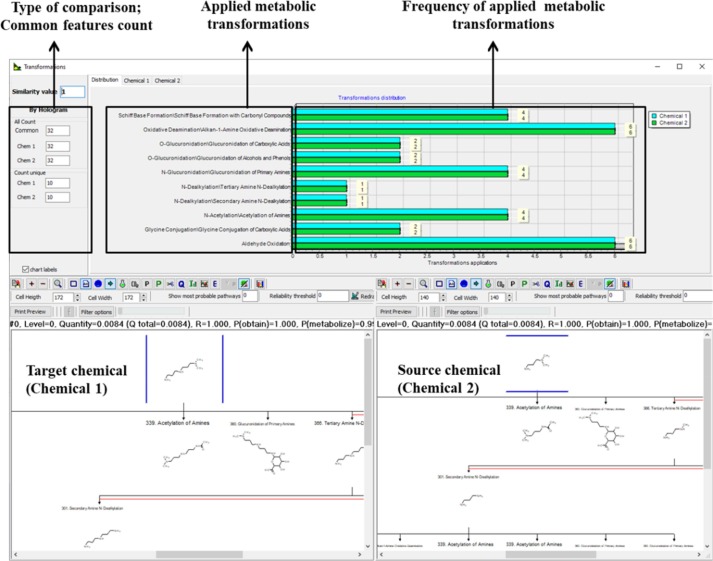
Assessing the commonality of the metabolic transformations or enzyme-mediated reactions of the compared chemicals characterises the extent to which the chemicals follow the same metabolic pathways.
Example for metabolic similarity between two chemicals with respect to the simulated skin metabolism transformations:
- Metabolic similarity with respect to the common mechanistic or structural pattern
This similarity criterion uses existing profiling schemes available in QSAR Toolbox via the direct connection between OASIS software and QSAR Toolbox.
In this respect, the similarity could be based on commonality in:
- mechanisms - using endpoint-specific profilers (e.g. Protein binding for Skin sensitization by OASIS profiler) OR
- structural features - using structure-based profilers (e.g. Organic functional groups profiler)
Example for metabolic similarity between two chemicals with respect to the identified protein binding alerts for skin sensitization:
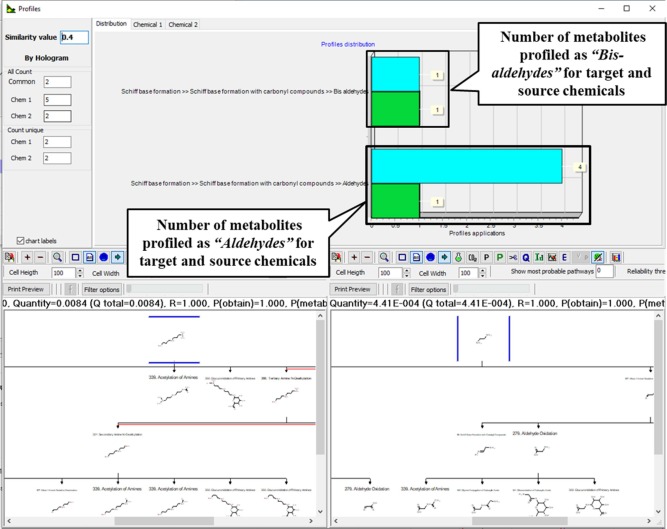
- Metabolic similarity with respect to the common metabolites
The third criterion, "Common metabolites", allows for the estimation of similarity based on the presence of common metabolites between compared chemicals. This similarity criterion is of practical use for analysis in the environmental fate models where the toxicity effects are associated with stable biodegradation product(s) that exceeded a defined threshold. Often, parent chemicals not structurally similar degrade to the same metabolite(s) that is associated with the toxic effects. The functionality allows defining a threshold for the quantity of the compared stable degradants.
Watch YouTube videos:
