CATALOGIC Soil
Endpoint
The biodegradation CATALOGIC Soil model simulates the catabolic pathways of chemicals in terrestrial aerobic conditions.
Data
The training set contains documented biodegradation pathways in soil for more than 300 chemicals, compiled into a searchable electronic database. The database contains 2302 metabolites for 318 chemicals. Microbial and metabolite information was collected from various sources, including articles and public web sites such as the database of European Food Safety Authorities [1].
Model
Simulation of metabolism is focused on correct reproduction of experimentally observed metabolites, only. The modeling was based on the set of principal molecular transformations and logic of metabolism as implemented in CATALOGIC 301C model [2, 3]. This set of transformations was expanded with an additional set of transformations extracted from the collected observed pathways in soil. The principal transformations were separated into groups with differing hierarchy. Further details on the mathematical formalism of the model can be reviewed in [4].
Domain
The stepwise approach [5] was used to define the applicability domain of the model. It consists of the following sub-domain levels:
- General parametric requirements - includes ranges of variation log KOW and MW,
- Structural domain - based on atom-centered fragments (ACFs).
A chemical is considered In Domain if its log KOW and MW are within the specified ranges and its ACFs are presented in the training chemicals. The information implemented in the applicability domain is extracted from the correctly predicted training chemicals used to build the model and in this respect the applicability domain determines practically the interpolation space of the model.
Performance
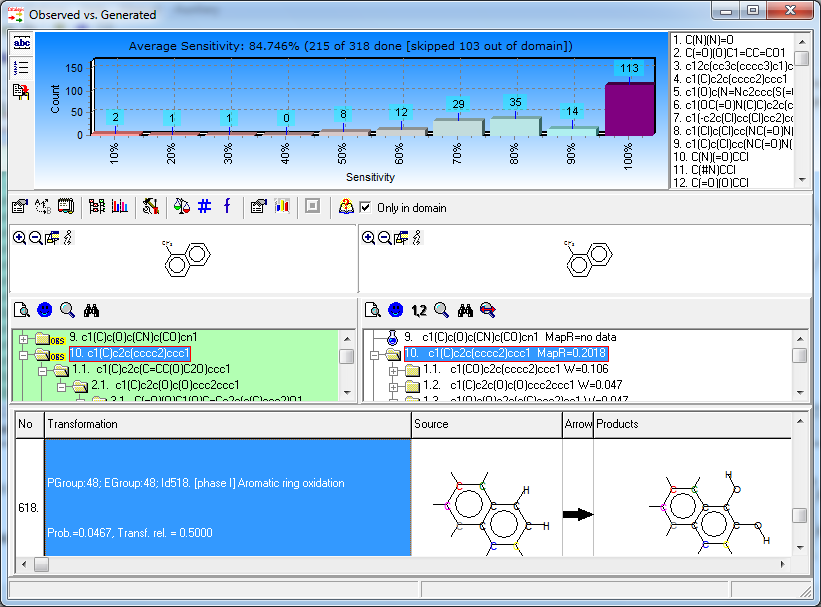
The similarity between generated and documented metabolic pathways was estimated by:
- Sensitivity - 85% (probability that the metabolite is predicted, given that the metabolite is truly observed)
- Predictability - 58% (probability that the metabolite is truly observed, given that the metabolite is predicted).
Reporting
The model provides results for:
- Quantities of parent and biodegradation products, mol/mol parent,
- Applicability domain details.
References
1. EFSA, European Food Safety Authorities Available at: http://dar.efsa.europa.eu/dar-web/provision.
2. S Dimitrov, T Pavlov, G Veith, O Mekenyan. SAR and QSAR
in Environ Res, 22, 2011, 699-718.
3. S Dimitrov, T Pavlov, N Dimitrova, D Georgieva, D Nedelcheva, A
Kesova, R Vasilev, O Mekenyan. SAR and QSAR in Environ Res, 22,
2011, 719-755.
4. S. Dimitrov, D. Nedelcheva , N. Dimitrova and O. Mekenyan. Sci
Total Environ. 408(18), 2010, 3811-3816.
5. S. Dimitrov, G. Dimitrova, T. Pavlov, N. Dimitrova, G.
Patlevisz, J. Niemela and O. Mekenyan, J. Chem. Inf. Model. 45
(2005), pp. 839-849.