Tetrahymena pyriformis IGC50 48h
Endpoint
The acute aquatic toxicity model predicts the concentration of a substance that inhibits 50% of the growth (IGC50) of the test population Tetrahymena pyriformis within a designated period.
Data
The training set consists of IGC50 values for 1330 chemicals [1-17]:
- Test duration - 48 hours,
- Test species - Tetrahymena pyriformis (ciliate protozoa).
The tested chemicals belong to the following categories:
- Narcotic toxicants - 351 chemicals,
- Phenols and anilines - 353 chemicals,
- Narcotic amines - 30 chemicals,
- Esters - 101 chemicals,
- Aldehydes - 94 chemicals,
- α, β- unsaturated alcohols - 32 chemicals,
- Reactive unspecified chemicals - 304 chemicals.
Model
The organism response to the presence of toxicant in the environment is considered as a consequence of the combined influence of two different processes: uptake of the chemical into the biophase and interaction with the site of action [18]. In the present model, the uptake is modeled by maximum potential of the toxicant to bioconcentrate in the fish, while the interaction of chemicals is explained by descriptors assessing the electrophilic character of the molecule [19]. Such descriptors could include the energy of the lowest unoccupied molecular orbital, electronegativity, average or maximum superdelocalizability, maximum charge at non-hydrogen atom, etc. The following models were developed based on regression analysis of the data:
Narcotic toxicants
log 1/IGC50 = 1.44(±0.04)+1.00(±0.02)logBCFmax -0.15(±0.01) ELUMO
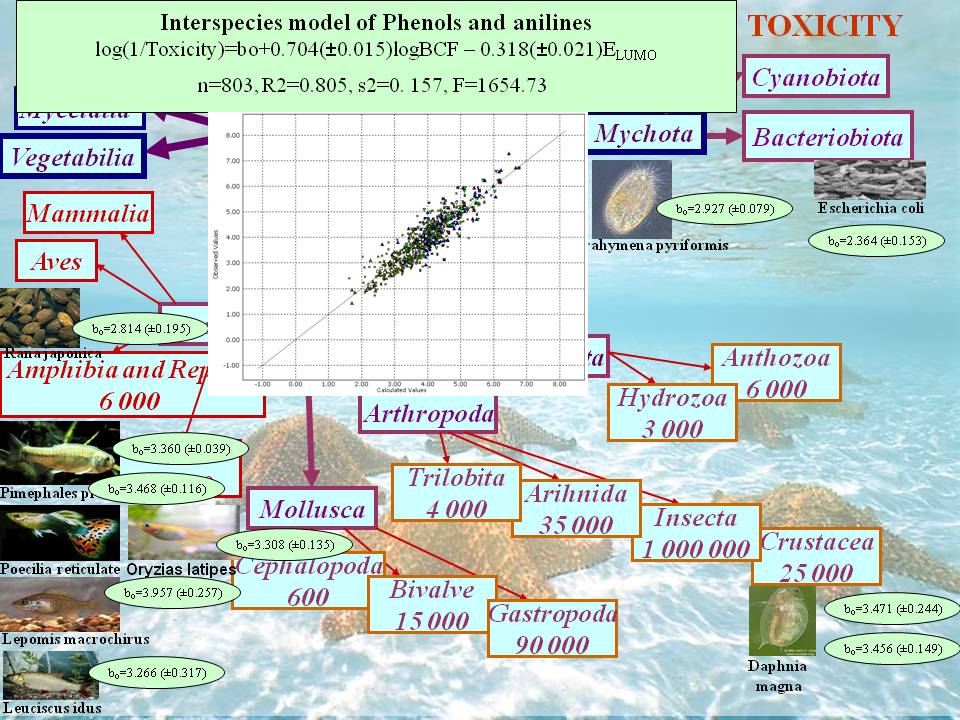
Phenols and anilines
log 1/IGC50 = 2.17(±0.05)+0.78(±0.03)logBCFmax -0.39(±0.03) ELUMO
Narcotic amines
log 1/IGC50 = 2.05(±0.11)+0.79(±0.08)logBCFmax
Esters
log 1/IGC50 = 1.30(±0.09)+1.05(±0.04)logBCFmax -0.39(±0.04) ELUMO
Aldehydes
log 1/IGC50 = -1.61(±1.21)+0.67(±0.06)logBCFmax +13.51(±4.03) ODDI
α, β- Unsaturated alcohols
log 1/IGC50 = 1.82(±0.13)+0.91(±0.09)logBCFmax -15.97(±4.84) ELUMO
where BCFMax is the maximum bioconcentration factor [20], ELUMO is the energy of the lowest unoccupied molecular orbital, ODDI is the donor delocalizability indices of the aldehyde O-atom and QC is the charge of the C atom from α, β- unsaturated alcohols. For the reactive unspecified chemicals, only the minimum toxicity is determined based on the model for narcotic chemicals (i.e., of narcotics).
Domain
The stepwise approach [21] was used to define the applicability domain of the model. It consists of the following sub-domain levels:
- General parametric requirements - includes ranges of variation of log KOW and MW,
- Structural domain - based on atom-centered fragments (ACFs).
A chemical is considered In Domain if its log KOW and MW are within the specified ranges and its ACFs are presented in the training chemicals. The information implemented in the applicability domain is extracted from the correctly predicted training chemicals used to build the model and in this respect the applicability domain determines practically the interpolation space of the model.
Statistics
The precision of the regression models is characterized by the following estimates: the 95% confidence intervals of model parameters, coefficient of determination (R2), mean squared error (estimate of error variance, s2), F value:
Narcotic toxicants
- Coefficient of determination R2 = 0.90,
- Mean squared error (estimate of error variance) s2 = 0.10,
- F value = 1579.39,
- Number of chemicals, n = 351.
Phenols and anilines
- Coefficient of determination R2 = 0.77,
- Mean squared error (estimate of error variance) s2 = 0.15,
- F value = 574.8,
- Number of chemicals, n = 353.
Narcotic amines
- Coefficient of determination R2 = 0.77,
- Mean squared error (estimate of error variance) s2 = 0.19,
- F value = 92.18,
- Number of chemicals, n = 30.
Esters
- Coefficient of determination R2 = 0.89,
- Mean squared error (estimate of error variance) s2 = 0.09,
- F value = 412.7,
- Number of chemicals, n = 101.
Aldehydes
- Coefficient of determination R2 = 0.61,
- Mean squared error (estimate of error variance) s2 = 0.16,
- F value = 71.90,
- Number of chemicals, n = 94.
α, β- Unsaturated alcohols
- Coefficient of determination R2 = 0.81,
- Mean squared error (estimate of error variance) s2 = 0.22,
- F value = 62.44,
- Number of chemicals, n = 32.
References
1. Schultz TW, Bryant SE, Lin DT. 1994. Bull Environ Contam
Toxicol 52:279-285.
2. Schultz TW, Cronin MTD. 1999. J Chem Inf Comput Sci
39:304-309.
3. Schultz TW. 1997. Toxicol Methods 7:289-309.
4. Schultz TW, Cronin MTD. 1997. Toxicol. Chemistry 16:
357-360.
5. Schultz TW, Sinks GD, Cronin MTD. 1997 Aquatic Toxicology 39:
267-278.
6. Cronin MTD, Bowers GS, Sinks GD, Schultz TW. 2000. SAR/QSAR
Environ. Research. 11: 301-312.
7. Bearden AP, Schultz TW. 1997. Environ. Toxicol. Chemistry 16:
1311-1317.
8.Schultz TW, Lin DT, Arnold LM. 1991. The Science of the Total
Environment 109/110: 569-580.
9. Schultz, TW, Mekenyan OG. 2005. Response-surface analyses: An
overview of an approach to predicting acute toxicity. In: Walker,
J.D. (ed), QSARs for Predicting Ecological Effects of Chemicals.
SETAC Press, Pensacola, FL, USA, pp. (in press).
10. Seward JR, Schultz TW. 1999. SAR/QSAR Environ. Research. 10:
557-567.
11. Schultz TW, Wilke TS, Bryant SE, Hosein LM. 1991. The Science
of the Total Environment 109/110: 581-587.
12. Cronin MTD, Gregory BW, Schultz, TW.1998. Chem. Res. Toxicol.
11: 902-908.
13. Cronin, M.T.D., Aptula, A.O., Duffy, J.C., Netzeva, T.I.,
Rowe, P.H., Valkova, I.V., 14. Schultz, T.W. 2002. Chemosphere 49:
1201-1221.
14. Schultz TW, Lin DT, Wilke TS, Arnold LM. QSAR for the
Tetrahymena pyriformis population growth endpoint: a mechanism of
action approach. In: Practical Applications of QSAR in
Environmental Chemistry and Toxicology (Devillers J, Karcher W,
eds). Dordrecht:Kluwer Academic Publishers, 1996;241-262.
15. Akers KS, Sinks GD, Schultz TW. 1999. Environmental Toxicology
and Pharmacology 7: 33-39.
16. Jaworska JS, Hunter RS, Gabble JR, Schultz TW.
Structure-Activity relationships for Diesters:Aquatic Toxicity to
Tetrahymena. In: Quantitative Structure-Activity relationships in
Environmental Sciences-VII (Fei Chen, Schuurmann, eds) SETAC
Special Publications Series, 1997.
17. Schultz TW, Sinks GD, Cronin MTD. Identification of Mechanisms
of Toxic Action of Phenols to Tetrahymena pyriformis from Molecular
Descriptors. In: Quantitative Structure-Activity relationships in
Environmental Sciences-VII (Fei Chen, Schuurmann, eds) SETAC
Special Publications Series, 1997.
18. J.W. McFarland, J. Med. Chem. 13 (1970) 1092-1196.
19. S.D. Dimitrov, O.G. Mekenyan, G.D. Sinks, T.W. Schultz,
Journal of Molecular Structure (Theochem, 622 (2003) 63-70.
20. S. Dimitrov, N. Dimitrova, D. Georgieva, K. Vasilev, T.
Hatfield, J. Straka, O. Mekenyan, SAR and QSAR in Environmental
Research, 23 (2012) 17-36.
21. S. Dimitrov, G. Dimitrova, T. Pavlov, N. Dimitrova, G.
Patlevisz, J. Niemela and O. Mekenyan, J. Chem. Inf. Model. 45
(2005) 839-849.