Chromosomal aberration
Endpoint
In vitro chromosomal aberrations (CA) test identifies agents that cause structural chromosome aberrations in cultured mammalian cells [1]. Chromosomal aberrations are changes affecting chromosome morphology. In general, CA involves breaking of chromosome segments, their loss or union with same or different chromosomes.
Data
Collected are 1077 chemicals with documented CA data without accounting for S9 metabolic activation [2]. Of them, positive as parents are 438 chemicals, whereas 639 chemicals are CA negative. Chemicals which are positive as parents have been used for deriving the alerts (see next section). CA data obtained after S9 metabolic activation have been collected for another set of chemicals (397). Of them, positive are 99 chemicals (which are negative as parents) and 298 chemicals are CA negative (as parents and metabolites). These two types of training sets are considered as biologically dissimilar in the modeling process; i.e., chemicals being mutagenic as parents are distinguished from chemicals, which are mutagenic after metabolic activation. These subsets are subsequently modeled separately by the TIMES structural alerts approach. In this respect, there are two CA models: with and without S9 metabolic activation included in the TIMES system.
Alerts in CA models
Direct binding of chemicals to DNA
is one of the underlying mechanisms that is responsible for CA
mutagenicity [3]. Disturbance of protein synthesis due to
inhibition of topoisomerases and interaction of chemicals with
nuclear proteins associated with DNA (e.g., histone proteins) are
identified as additional mechanisms also leading to positive CA
effect. Based on that, the toxicodynamic component of the CA
models, i.e. alerts (functionalities responsible for eliciting CA)
are derived to account for interactions with DNA and/or proteins.
Alerts are developed by chemicals which are positive as parents
(one should be reminded that chemicals which are positive only
after metabolic activation are negative as parents). Alerts
associated with the CA models consist of boundaries, which provide
the minimum structural requirements for interacting with DNA and/or
proteins. Usually, additional structural and parametric boundaries
are required for completing definition of alerts.
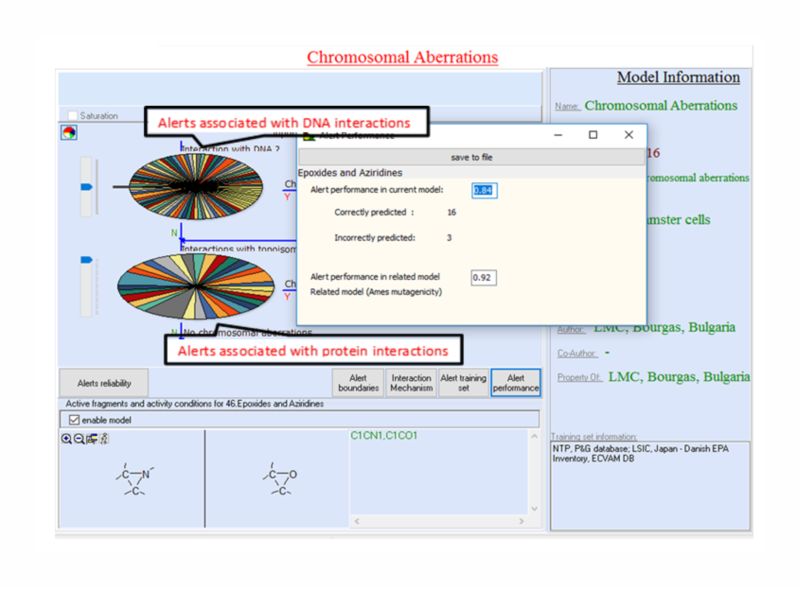
A subset of the training set is associated with each alert. In
fact, these are chemicals used to define alerts boundaries.
Cardinality of these local training sets depends on the number of
substances captured by the alert boundaries. Only those alerts are
included in the model for which a clear interpretation of the
molecular interaction mechanism could be provided. Currently, the
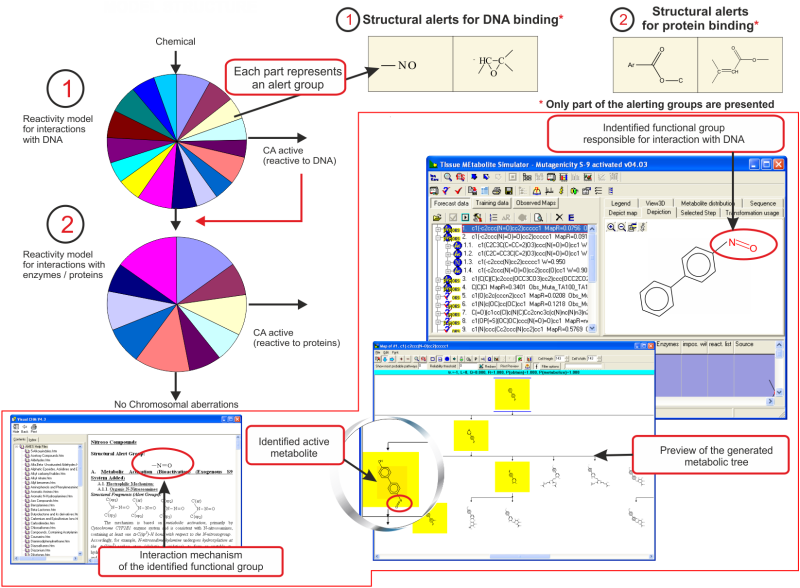
total number of alerting groups included in the TIMES CA models is
124, where 91 of them are associated with DNA binding and 33 are
associated with protein binding [4].
CA model without accounting for metabolic activation (TIMES_CA (-S9))
The training set of the CA model (-S9) comprises chemicals which are positive (438) and negative (639) as parents. In this respect, the model predicts DNA and/or protein damaging effect of parent chemicals, only. Positive predictions are obtained when the complete set of structural/parametric boundaries of the alert(s) are met in the parent chemicals.
According to the performance of TIMES_CA model (-S9) when applied on the training set chemicals, correct predictions are provided for:
• 362 out of 438 observed positive parents (Sensitivity =
83%)
• 580 out of 639 observed negative parents (Specificity =
91%)
CA model accounting for metabolic activation (TIMES_CA (+S9))
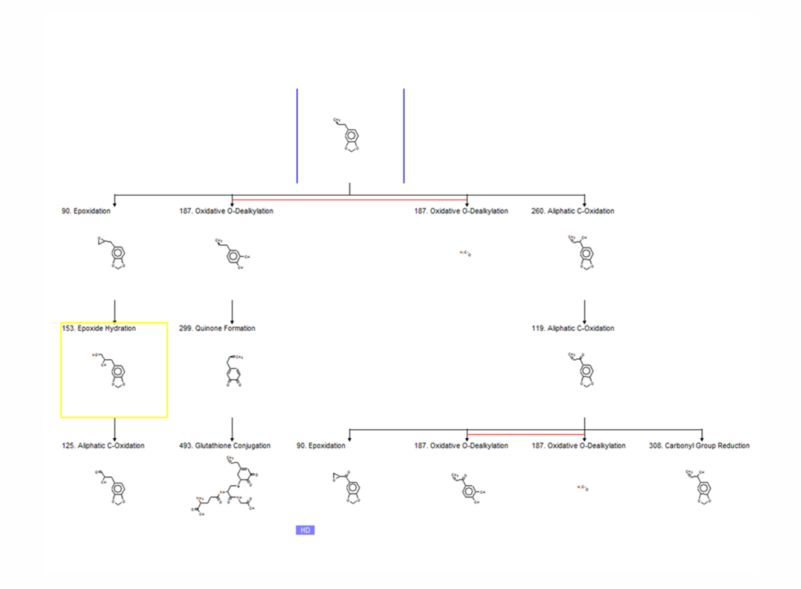
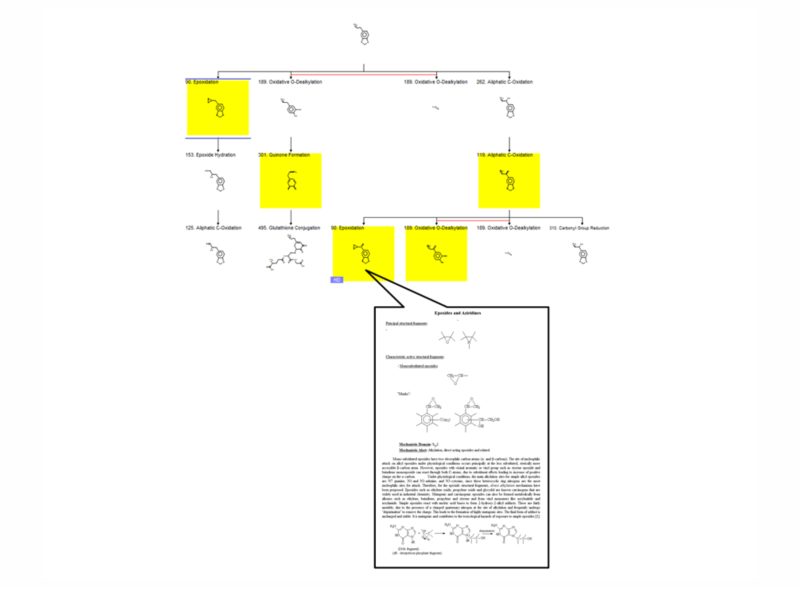
The training set of the CA model (+S9) comprises substances which are positive (99) and negative (298) after metabolic activation. Since both CA models (-S9 and +S9) share the same alerts, the CA model (+S9) predicts DNA and/or protein damaging effect of parent chemicals and their metabolic products, obtained after S9 metabolic activation. The toxicokinetic component of the model consists of a metabolic simulator which is trained to reproduce documented maps for mammalian liver metabolism of 261 chemicals. The number of generated metabolites is manageable which allows combining in a single modeling platform metabolic simulation of chemicals and their interaction with target macromolecules. The S9 metabolic simulator has its own training set which includes 542 metabolic reactions:
• Phase I transformations, such as aliphatic C-oxidation,
aromatic C-hydroxylation, oxidative N- and O-dealkylation,
epoxidation, ester and amide hydrolysis, carbonyl group reduction,
nitro group reduction, N-hydroxylation, etc.
• Phase II transformations, such as glucuronidation,
sulfation, glutathione conjugation, N-acetylation, etc.
A characteristic feature of the in vitro metabolic simulator is the significant prevalence of Phase I reactions (85-90%) over Phase II reactions, which reflects the specificity of the in vitro experimental systems with respect to their metabolic capacity [5]. The metabolic simulator cannot estimate the reaction rates (e.g. kinetic constants, half-lives, etc.) but is able to provide tentative information for their probability. The logic of the in vitro CA model (+S9) is illustrated in Figure 1:
Figure 1. Illustration of the integral CA mutagenicity prediction based on metabolic simulator and defined structure-activity relationships (SARs).
Parent chemicals and each of the generated metabolites are submitted to a battery of SARs (alerts) to screen for a general CA effect and mutagenicity mechanisms. Thus, chemicals are predicted to be mutagenic as parents only, parents and metabolites, and metabolites only.
According to the performance of TIMES_CA model (+S9) when applied on the training set chemicals, correct predictions are provided for:
• 71 out of 99 observed positive after metabolic activation
(Sensitivity = 72%)
• 223 out of 298 observed negative after metabolic activation
(Specificity = 75%)
Domain
Each of the TIMES CA models (-S9 and +S9) has its own model
applicability domain, which supports the CA predictions [6]. The
applicability domains consist of three sub-domain layers: general
parametric requirements, structural features and alert(s)
reliability. Two chemical subsets are used for deriving the model
domains. The first subset includes the training set chemicals which
are correctly predicted by the models, whereas the second subset
comprises training set chemicals which are incorrectly predicted by
the models.
The correct chemical subset is used for defining the general
parametric requirements. Extracted are specific ranges of the
molecular weight (MW) and the 1-octanol/water partition coefficient
(log KOW):
• Molecular weight MW (in Da) ϵ [44; 1,416],
• log KOW ϵ [-10, 30].
The atom-centered fragments extracted from the correct subset of
chemicals are used to define the structural domain. Briefly, the
structural domain is assessed based on atom-centered fragments,
extracted from correctly and incorrectly predicted (i.e., false
positives and false negatives) substances from the model training
sets by accounting for the atom type, hybridization and attached
H-atoms of the central atom and its first neighbours. If the
neighbour is a heteroatom then the diameter of the fragment is
increased up to three consecutive heteroatoms or to the first
carbon atoms in sp3 hybridization. In order to assess if a new
chemical belongs to the structural domain, the system partitions
the chemical to atom-centered fragments, which are then matched to
the fragments extracted from the correct and incorrect chemical
subsets. The new chemical is estimated to belong to the structural
domain only when its atom-centered fragments are found in the list
of correct fragments.
The third level of the domain accounts for reliability of alerts. Currently, information for alerts reliability is provided in the model reports.
Reporting
The predictions of in vitro Chromosomal aberrations models could be reported as:
• A tab delimited text file providing the following
information for chemical: identity (CAS number, Name, SMILES),
observed CA mutagenicity, predicted CA mutagenicity, information
for the alert(s) responsible for DNA and/or protein interaction,
alert reliability and performance, applicability domain, etc.
• QSAR Prediction Reporting Format (QPRF) is available,
generating a report for one chemical or group of chemicals.
The TIMES CA (+S9) model provides information for:
• Simulated metabolic map,
• Positive parents and/or metabolites,
• Mechanisms of DNA and/or protein damage,
• Local training set of alerts,
• Performance of alerts,
• Different types of reporting
• …..
Decision support character of TIMES_CA models
Users of the TIMES model may assess reliability of the predictions by evaluating the amount of mechanistic justification supporting the ultimate prediction. In this respect, the modeling results could be considered as decision supporting rather than decision making outcome.
References
1. Evans,
H.J. 1976. Cytological Methods for Detecting Chemical Mutagens. In:
Chemical Mutagens, Principles and Methods for their Detection, Vol.
4, Hollaender, A. (ed) Plenum Press, New York and London, pp.
1-29.
2. Sofuni, T., Ed. 1998. Data Book of
Chromosomal Aberration Test in vitro, Revised Edition. Life-Science
Information Center, Tokyo, Japan.
3. Mekenyan, О., Todorov, М., Serafimova,
Р., Stoeva, С., Aptula, А., Finking, Р., Jacob, Е. 2007.
Identifying the structural requirements for chromosomal aberration
by incorporating molecular flexibility and metabolic activation of
chemicals. Chem. Res. Toxicol. Vol. 20, pp. 1927−1941.
4. Petkov, P.I., Schultz, T.W., Donner,
E.M., Honma, M., Morita, T., Hamada, Sh., Wakata, A., Mishima, M.,
Maniwa, J., Todorov, M., Kaloyanova, E., Kotov, S., Mekanyan, O.
2016. Integrated approach to testing and assessment for predicting
rodent genotoxic carcinogenicity, J. Appl. Toxicol. 36(12):
1536-1550.
5. Mekenyan, O., Dimitrov, S., Pavlov, T.,
Dimitrova, G., Todorov, M., Petkov, P., Kotov, S. 2012. Simulation
of chemical metabolism for fate and hazard assessment. V. Mammalian
hazard assessment, SAR and QSAR in Environmental Research, 23:5-6,
553-606.
6. Dimitrov, S., Dimitrova, G., Pavlov, T.,
Dimitrova, N., Patlewicz, G., Niemela J., Mekenyan, O. 2005. J.
Chem. Inf. Model., Vol. 45, pp. 839-849. .