in vivo Liver Clastogenicity
Endpoint
Clastogenicity detects substances that can cause breaks in chromosomes leading to sections of the chromosomes being deleted, added or rearranged [1]. Since the liver is the main organ that metabolizes chemicals, in vivo liver micronucleus/chromosomal aberrations tests are assumed to be sufficient for detection of clastogens as parents and after metabolic activation in liver [2].
Data
Training set of the model consists of 115 chemicals with integrated liver clastogenicity data from two in vivo liver mutagenicity tests: micronucleus test (107 chemicals) and chromosomal aberration test (8 chemicals). 78 of 115 chemicals show positive response and 37 chemicals are non-clastogenic. The TIMES liver clastogenicity model consists of three components: alerts, metabolic activation and metabolic detoxification of substances.
Alerts in the liver clastogenicity model
The working hypothesis used to develop each of the in vivo TIMES mutagenicity models is that reactivity of chemicals in in vitro and in in vivo environment is the same. The only difference is bioavailability, i.e. distribution of reactive species in systematic circulation (see next sections). Reactivity of chemicals towards DNA and/or proteins are the underlying mechanisms that are responsible for clastogenicity [3]. In this respect, the toxicodynamic component of the liver clastogenicity model, i.e. alerts (functionalities responsible for eliciting mutagenicity) is assumed to be the same as in the in vitro chromosomal aberrations model, i.e. accounting for interactions with DNA and/or proteins. 91 alerts accounting for DNA damage and 33 alerts accounting for protein damage have been derived. Alerts consist of two layers of structural boundaries. First, minimum structural requirements for interacting with DNA/proteins have been developed. Second, additional structural/parametric boundaries have been added for completing definition of alerts. While minimum structural requirements could bring positive and negative effect, the additional structural/parametric boundaries provide the sufficient conditions for positive effect only.
A subset of the training set is associated with each alert. In fact, these are the chemicals captured by the minimum structural requirements of alerts. The TIMES system provides local training sets of the clastogenicity model as well as the local subset of chemicals associated with same alert in the in vitro chromosomal aberrations model.
Metabolic activation in the liver clastogenicity model
The in vivo metabolic simulator reproduces the multi-pathway xenobiotic metabolism in living rats on the basis of metabolic pathways of xenobiotic chemicals [4]. Current version of the simulator contains 506 structurally generalized molecular transformations, which were subdivided into:
- 26 abiotic (non-enzymatic) transformations (e.g. tautomerization, acyl halide hydrolysis, geminal diol dehydration, etc.), which occur mostly spontaneously,
- 415 Phase I enzymatic transformations (e.g. aliphatic C-oxidation, epoxidation, aromatic C-hydroxylation, ester hydrolysis, amide hydrolysis, dehalogenation, etc.),
- 65 Phase II enzymatic transformations (e.g. glucuronidation, glutathione conjugation, sulfation, acetylation, etc.).
Metabolic transformations are hierarchically prioritized based on their feasibility of occurrence. Feasibility estimates are directly associated with formal kinetic constants. Non-enzymatic transformations have the highest feasibility of occurrence, followed by enzymatic transformations. Metabolic activation of parent chemicals is illustrated in Figure 1:
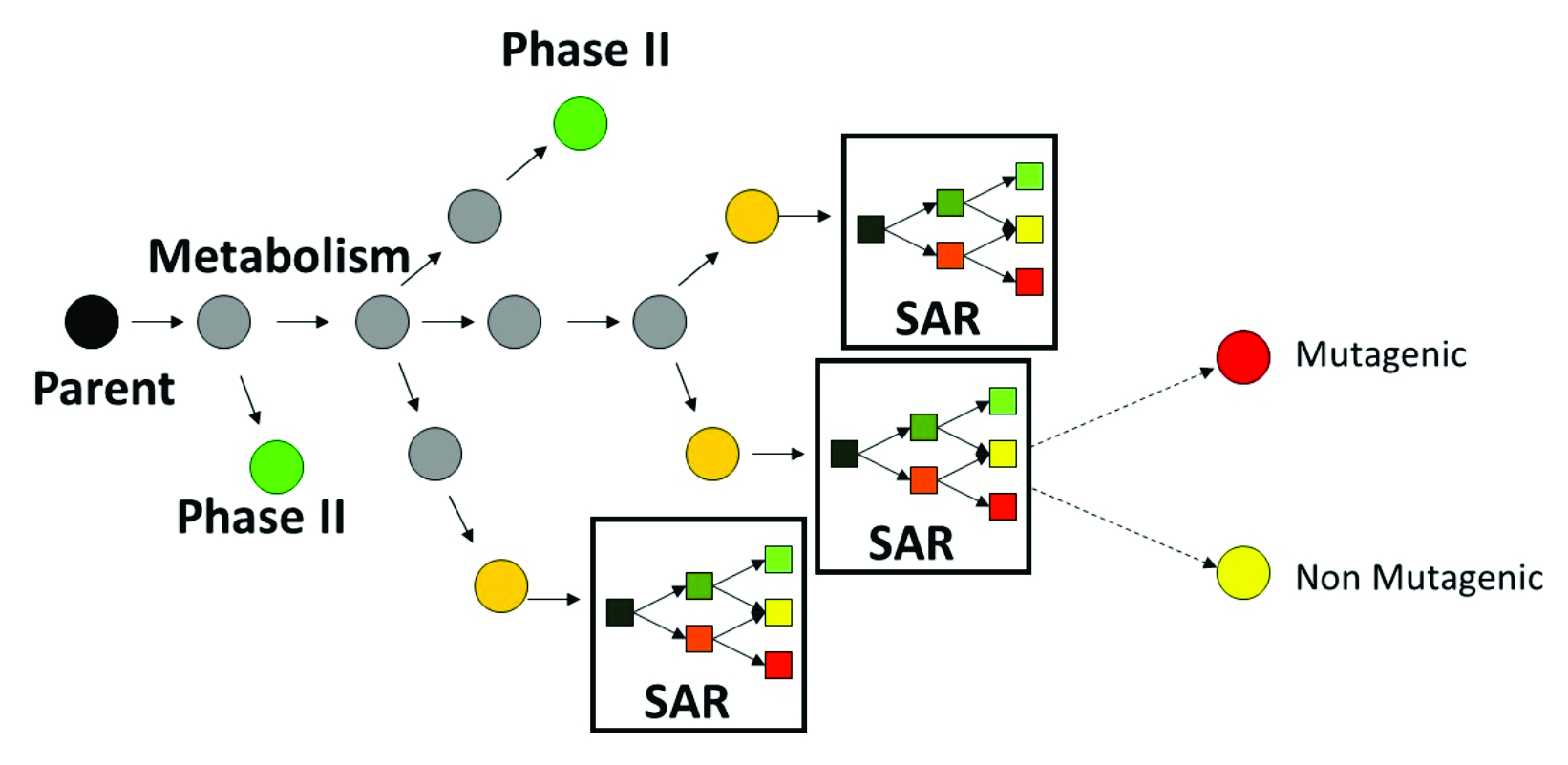
Figure 1. Metabolic activation of parent chemicals
Metabolic detoxification in the liver clastogenicity model
Implementation of metabolic detoxification pathways in the in vivo model has been introduced to predict mutagenicity particularly in liver as the main organ for xenobiotic metabolism.
In in vitro environment, all generated metabolites are theoretically available to interact (almost stochastically) with macromolecules present in the incubation medium, and thus have the potential to elicit a mutagenicity effect [4]. In in vivo environment, enzymes are aggregated in multienzyme complexes and the cells could be protected from reactive metabolites via shuttling intermediates between consecutive enzymes. The channelling effect allows the product of one enzymatic reaction to become a substrate of the subsequent enzymatic reaction. It can be therefore assumed that the in vitro active chemicals and/or their active metabolites are not freely available in vivo to cause damage. The majority of these metabolites are considered to be "trapped" across the in vivo detoxification pathways. For some in vitro mutagens (e.g. aromatic amines possessing polar functional groups) it has been shown that the parent chemicals and/or their metabolites could be "trapped" in liver detoxification pathways, as shown in Figure 2.
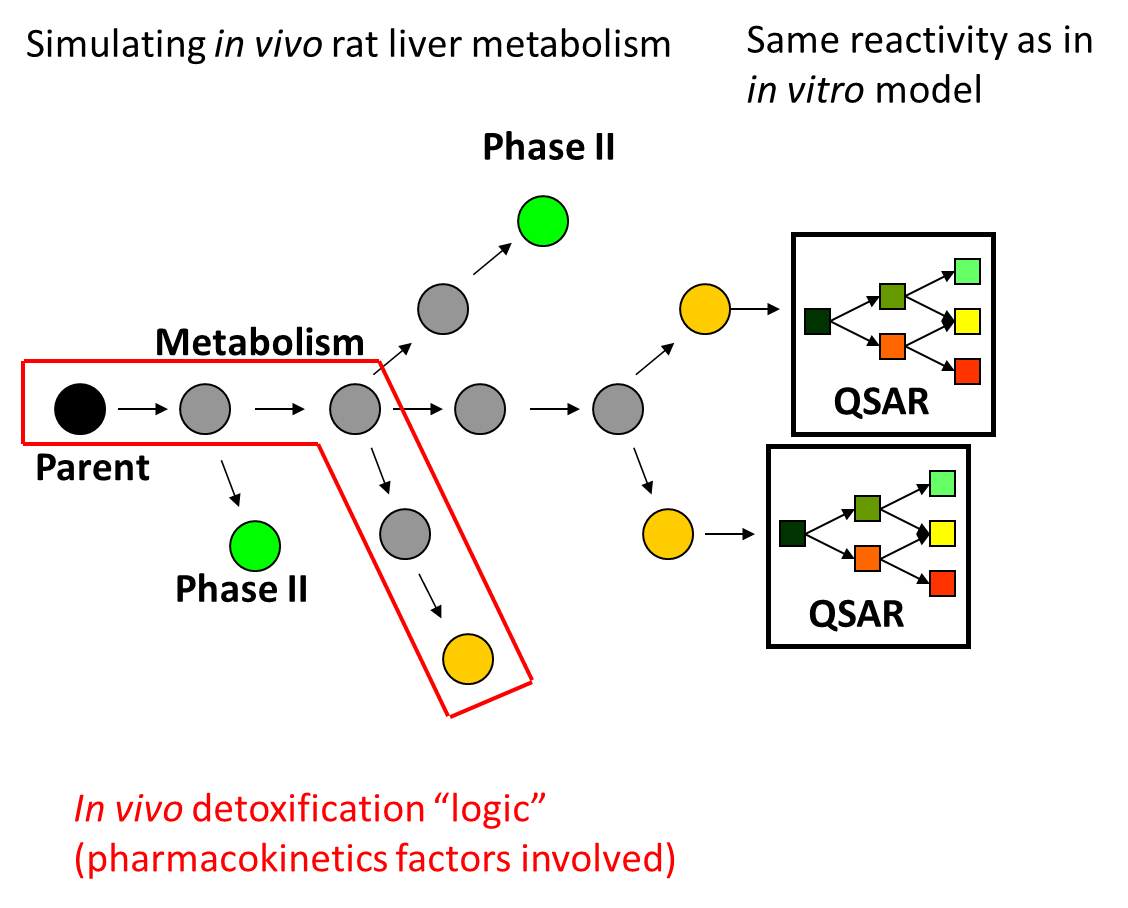
Figure 2. Application of metabolic detoxification pathways on parent chemicals and their generated metabolites.
Parent chemicals and each of the generated metabolites are submitted to a battery of alerts (SARs) to screen for a general DNA and/or protein binding and mutagenicity mechanisms. Subsequently, a list of predefined in vivo metabolic detoxification pathways has been applied on parent chemicals and their metabolites. Only these parents and metabolites, which are not involved in "trapping" detoxification pathways and are captures by alerts containing the complete set of structural boundaries are predicted to be clastogenic as parents, parents and metabolites, and metabolites only.
According to the performance of in vivo TIMES liver clastogenicity model when applied on the training set chemicals, correct predictions are provided for:
- 62 out of 78 observed positive (Sensitivity = 79%)
- 27 out of 37 observed negative (Specificity = 73%)
Domain
Applicability domain of the liver clastogenicity model consists of three sub-domain layers: general parametric requirements, structural features and alert(s) reliability [5]. Two chemical subsets are used for deriving the model domains. The first subset includes the training set chemicals which are correctly predicted by the models, whereas the second subset comprises training set chemicals which are incorrectly predicted by the models.
The correct chemical subset is used for defining the general parametric requirements. Extracted are specific ranges of the molecular weight (MW) and the 1-octanol/water partition coefficient (log KOW):
• Molecular weight MW (in Da) ϵ [58; 698],
• log KOW ϵ [-3, 13].
The atom-centered fragments extracted from the correct subset of chemicals are used to define the structural domain. Briefly, the structural domain is assessed based on atom-centered fragments, extracted from correctly and incorrectly predicted (i.e., false positives and false negatives) substances from the model training sets by accounting for the atom type, hybridization and attached H-atoms of the central atom and its first neighbours. If the neighbour is a heteroatom then the diameter of the fragment is increased up to three consecutive heteroatoms or to the first carbon atoms in sp3 hybridization. In order to assess if a new chemical belongs to the structural domain, the system partitions the chemical to atom-centered fragments, which are then matched to the fragments extracted from the correct and incorrect chemical subsets. The new chemical is estimated to belong to the structural domain only when its atom-centered fragments are found in the list of correct fragments.
The third level of the domain accounts for reliability of alerts. Currently, information for alerts reliability is provided in the model reports.
Reporting
The predictions of in vivo liverclastogenicity model could be reported as:
- A tab delimited text file providing the following information for substances: identity (CAS, Name, SMILES), documented liver clastogenicity, predicted liver clastogenicity, information for the alert(s) responsible for DNA/protein interaction, alert reliability and performance, applications of detoxification pathways, applicability domain, etc.
- QSAR Prediction Reporting Format (QPRF) is available, generating a report for one chemical or group of chemicals.
Decision support character of TIMES liver clastogenicity model
Users of the TIMES model may assess reliability of the predictions by evaluating the amount of mechanistic justification supporting the ultimate prediction. In this respect, the modeling results could be considered as decision supporting rather than decision making outcome.
References
- Rosefort C., Fauth E., Zankl H. 2004. Micronuclei induced by aneugens and clastogens in mononucleate and binucleate cells using cytokinesis block assay. Mutagenesis 19, pp. 277-284.
- Uno Y., Morita T., Luijten M., Beevers C., Hamada S., Itoh S., Ohyama W., Takasawa H. 2015. Recommended protocols for the liver micronucleus test: Report of the IWGT working group. Mutat Res Genet Toxicol Environ Mutagen, pp. 783:13-8.
- Petkov, P.I., Schultz, T.W., Donner, E.M., Honma, M., Morita, T., Hamada, Sh., Wakata, A., Mishima, M., Maniwa, J., Todorov, M., Kaloyanova, E., Kotov, S., Mekanyan, O. 2016. Integrated approach to testing and assessment for predicting rodent genotoxic carcinogenicity, J. Appl. Toxicol. 36(12): 1536-1550.
- Mekenyan, O., Dimitrov, S., Pavlov, T., Dimitrova, G., Todorov, M., Petkov, P., Kotov, S. 2012. Simulation of chemical metabolism for fate and hazard assessment. V. Mammalian hazard assessment, SAR and QSAR in Environmental Research, 23:5-6, 553-606.
- Dimitrov, S., Dimitrova, G., Pavlov, T., Dimitrova, N., Patlewicz, G., Niemela J., Mekenyan, O. 2005. J. Chem. Inf. Model., Vol. 45, pp. 839-849.

