in vivo Liver Genotoxicity
Endpoint
In vivo liver genotoxicity model identifies chemicals that cause DNA and/or protein damage in liver of rats or mice.
Data
The training set consists of 251 chemicals [1,2] with integrated liver genotoxicity outcome from four different assays: Comet assay, unscheduled DNA synthesis, transgenic rodent mutation assay and liver micronucleus test. If one of the assays shows positive result, the ultimate liver genotoxicity assignation is considered to be positive.
Model
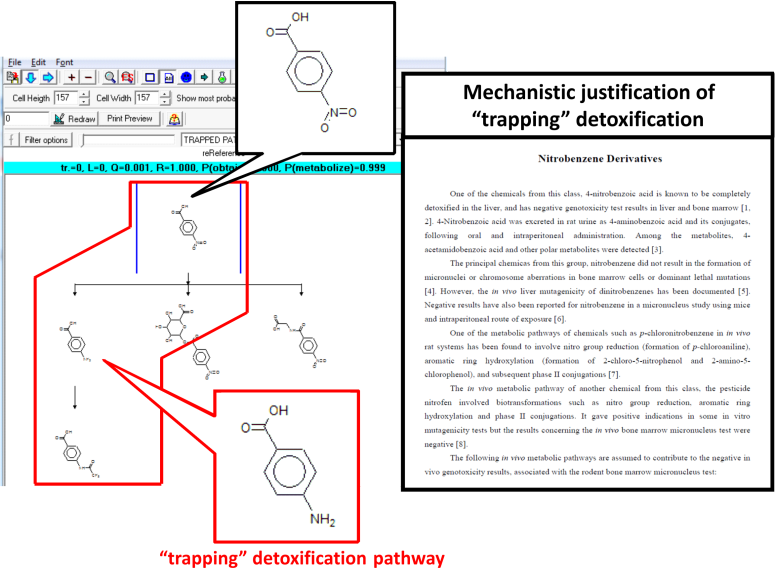
If availability of parent chemicals or their metabolites in the target tissue is not rate limiting, then hence no differences would be expected between the in vitro and in vivo results, i.e., the toxicodynamic model for in vitro should also be valid in vivo [3]. In this respect, in vitro reactivity taking into account interactions of chemicals with DNA and/or proteins should be suitable as part of the in vivo model for liver genotoxicity. Only those toxicophores having clear interpretation for the molecular mechanism causing the ultimate effect were included in the model. Some of the specified alerts interact directly with DNA or nuclear proteins, whereas others are applied in a combination of two-dimensional QSAR models assessing the degree of activation of the alerts from the rest of the molecules. In the in vivo liver genotoxicity model in vitro reactivity component is combined with in vivo metabolism simulator. The simulator was developed comprising a set of structurally generalized molecular transformations. A database of 220 in vivo metabolic pathways of chemicals was compiled and formed the training set used to derive the rat in vivo metabolic simulator. In vivo, enzymes are aggregated in multienzyme complexes and the cells could be protected from reactive metabolites via shuttling intermediates between consecutive enzymes. Thus, the product of one enzymatic reaction may become a substrate of the subsequent enzymatic reaction. In this so-called channeling effect, some in vitro positive metabolites could be "trapped" and thus unavailable to react with macromolecules in liver. In vitro positive chemicals which are not involved in "trapping" detoxification pathways are considered capable of causing DNA and/or protein damage and hence in vivo liver genotoxic effects. In vitro negative chemicals are also expected to be in vivo negative in liver. In vitro negative but in vivo liver positive genotoxicity results are observed for a very small number of chemicals only.
Domain
The stepwise approach [4] was used to define the applicability domain of the model. It consists of the following sub-domain levels:
- General parametric requirements - includes ranges of variation log KOW and MW,
- Structural domain - based on atom-centered fragments (ACFs).
- Interpolation space - estimates the population density of the parametric space defined by the explanatory variables of the QSAR models by making use the training set chemicals.
- Domain of simulator of metabolism - determines the reliability of the simulated metabolism.
A chemical is considered In Domain if its log KOW and MW are within the specified ranges and if its ACFs are presented in the training chemicals. The information implemented in the applicability domain is extracted from the correctly predicted training chemicals used to build the model and in this respect the applicability domain determines practically the interpolation space of the model.
Statistics
For 251 chemicals, the in vivo liver genotoxicity model was able to predict correctly 84% of the liver positive and 80% of the liver negative training set chemicals.
Reporting
The predictions from liver genotoxicity model could be reported as a tab delimited text file providing the following information for: chemical identity (CAS number, Name, SMILES), predicted in vitro CA mutagenicity, observed in vivo liver genotoxicity, predicted in vivo liver genotoxicity, information for the alert(s) responsible for DNA and/or protein interaction, "trapping" detoxification pathways, applicability domain, etc.
References
1. T. Morita, J.
MacGregor, M. Hayashi, Micronucleus assays in rodent tissues other
than bone marrow, Mutagenesis, Vol. 26, (2011), pp.
223-230.
2. M. Hayashi, J. MacGregor, D. Gatehouse, D.
Blakey, S. Dertinger, L. Abramsson-Zetterberg, G. Krishna, T.
Morita, A. Russo, N. Asano, H. Suzuki, W. Ohyama, D. Gibson, In
vivo erythrocyte micronucleus assay: III. Validation and regulatory
acceptance of automated scoring and the use of rat peripheral blood
reticulocytes, with discussion of non-hematopoietic target cells
and a single dose-level limit test Original Research Article
Mutation, Research Genetic Toxicology and Environmental
Mutagenesis, Vol. 627, (2007), pp. 10-30.
3. O. Mekenyan, P. Petkov, S. Kotov, S. Stoeva,
V. Kamenska, S. Dimitrov, M. Honma, M. Hayashi, R. Benigni, M.
Donner, G. Patlewicz, Investigating the Relationship between in
Vitro−in Vivo Genotoxicity: Derivation of Mechanistic QSAR Models
for in vivo liver genotoxicity and in vivo bone marrow micronucleus
formation which encompass metabolism, Chemical Research in
Toxicology, Vol. 25, (2012), pp. 277- 296.
4. S. Dimitrov, G. Dimitrova, T. Pavlov, N.
Dimitrova, G. Patlevisz, J. Niemela and O. Mekenyan, J. Chem.
Inf. Model. 45 (2005), pp. 839-849.