Mouse Lymphoma thymidine kinase (MLA) model
Endpoint
In vitro mouse lymphoma thymidine kinase assay (MLA-tk) identifies agents that cause point mutations, chromosomal rearrangement and aneuploidy [1,2]. The tk locus in mouse lymphoma cells is the preferred target for mammalian cell genotoxicity assays because of the wealth of data which exists to support this locus and assay and because of the broad spectrum of damages detected.
Data
Two types of training sets are used in the modeling process. The first training set consists of 460 chemicals with documented MLA-tk data without S9 metabolic activation separated in two subsets: 269 positive chemicals and 191 negative chemicals. The second training set consists of 313 chemicals having MLA data with S9 metabolic activation. Of them, 197 substances have positive data and 116 substances have negative data. These two types of training sets are considered as biologically dissimilar in the modeling process; i.e., chemicals being mutagenic as parents are distinguished from chemicals, which are mutagenic after metabolic activation. These subsets are subsequently modeled separately in TIssue MEtabolism Simulator (TIMES) system by definition of alerts (i.e. functionalities responsible for eliciting positive MLA effect).
The training set of the model for predicting MLA-tk without S9 activation is used for extracting alerting groups. On the other hand, the training set of the model for MLA-tk with S9 activation is used to adjust the simulator for S9 (rat liver mix) activation as well as to validate the already derived alerts. Thus, the combination of the S9 simulated metabolism and reactivity model (based on alert groups) applied to each generated metabolite has to reproduce successfully the metabolic activation of chemicals from this training set.
Data from a modified MLA procedure suggested that aneuploidy-related mechanisms are only manifested in a long-term treatment (24h) of cells [3]. Therefore, in the modelling process short-term and long-term MLA-tk data are combined to facilitate definition of alerts associated with aneuploidy.
Alerts in MLA model
Direct binding of chemicals to DNA is one of the underlying mechanisms that is responsible for MLA-tk mutagenicity. Disturbance of protein synthesis due to inhibition of topoisomerases and interaction of chemicals with nuclear proteins associated with DNA (e.g., histone proteins) are identified as additional mechanisms also leading to positive MLA-tk effect. Furthermore, MLA-tk system is capable of detecting compounds that induce aneuploidy. Specific feature of this model is that aneuploidy is modeled explicitly along with point mutation and clastogenicity. Aneuploidy is presumably associated with covalent and non-covalent tubulin binding reactions, whereas clastogenicity is related to interactions with other cellular proteins.
Fifty-two DNA and 36 protein binding alerts, supported by mechanistic justification, have been used in combination with a new in vitro rat liver S9 fraction metabolic simulator to evaluate MLA-tk mutagenicity. Five of these 36 protein binding alerts are explicitly associated with aneuploidy induction. Alerts are developed by chemicals which are positive as parents and consist of boundaries, which provide the minimum structural requirements for interacting with DNA and/or proteins. Usually, additional structural and parametric boundaries are required for completing definition of alerts.
A subset of the training set is associated with each alert. In fact, these are chemicals used to define alerts boundaries. Cardinality of these local training sets depends on the number of substances captured by the alert boundaries. Only those alerts are included in the model for which a clear interpretation of the molecular interaction mechanism could be provided.
MLA model without accounting for metabolic activation (TIMES_MLA-tk (-S9))
The training set of the MLA-tk model (-S9) comprises chemicals which are positive (269) and negative (191) as parents. In this respect, the model predicts DNA and/or protein damaging effect of parent chemicals, only. Positive predictions are obtained when the complete set of structural/parametric boundaries of the alert(s) are met in the parent chemicals.
According to the performance of TIMES_MLA-tk model (-S9) when applied on the training set chemicals, correct predictions are provided for:
- 224 predicted positive out of 269 parent structures observed to be positive give a sensitivity of 83%,
- 173 predicted negatives out of 191 parent structures observed to be negative give a specificity of 91%,
- 397 correct predictions out of the 460 measured parent structures give a concordance of 86%.
The latter result is consistent with the concordance of the MLA-tk tests with respect to its intra-laboratory repeatability (81%) and inter-laboratory reproducibility (91%) [4,5].
MLA model accounting for metabolic activation (TIMES_MLA-tk (+S9))
The training set of the MLA-tk model (+S9) comprises substances which are positive (197) and negative (116) after metabolic activation. Since both MLA models (-S9 and +S9) share the same alerts, the MLA-tk model (+S9) predicts DNA and/or protein damaging effect of parent chemicals and their metabolic products, obtained after S9 metabolic activation. The toxicokinetic component of the model consists of a metabolic simulator which is trained to reproduce documented maps for mammalian liver metabolism of 261 chemicals. The number of generated metabolites is manageable which allows combining in a single modeling platform metabolic simulation of chemicals and their interaction with target macromolecules. The S9 metabolic simulator has its own training set which includes 542 metabolic reactions:
- Phase I transformations, such as aliphatic C-oxidation, aromatic C-hydroxylation, oxidative N- and O-dealkylation, epoxidation, ester and amide hydrolysis, carbonyl group reduction, nitro group reduction, N-hydroxylation, etc.
- Phase II transformations, such as glucuronidation, sulfation, glutathione conjugation, N-acetylation, etc.
A characteristic feature of the in vitro metabolic simulator is the significant prevalence of Phase I reactions (85-90%) over Phase II reactions, which reflects the specificity of the in vitro experimental systems with respect to their metabolic capacity [6].
A probability of occurrence is ascribed to each principal transformation, which determines its hierarchy in the transformation list. The simulator starts by matching the parent molecule with the reaction fragment associated with the transformation having highest probability of occurrence. When a match is identified, the molecule is metabolized, and transformation product is treated as the parent molecules for the next metabolic step. The procedure is repeated for the newly formed metabolites until the product of probabilities of consecutively performed transformations reaches a user-defined threshold. This probability threshold is customized for different endpoints. For the TIMES_MLA-tk models this threshold is 0.1 and typically does not exceed this value for the other in vitro TIMES mutagenicity models.
When a new chemical is submitted for prediction, firstly, a manageable number of metabolites are generated. The parent chemical and each of the generated metabolites are submitted to the battery of alerting groups to screen for the effect, as shown in Figure 1:
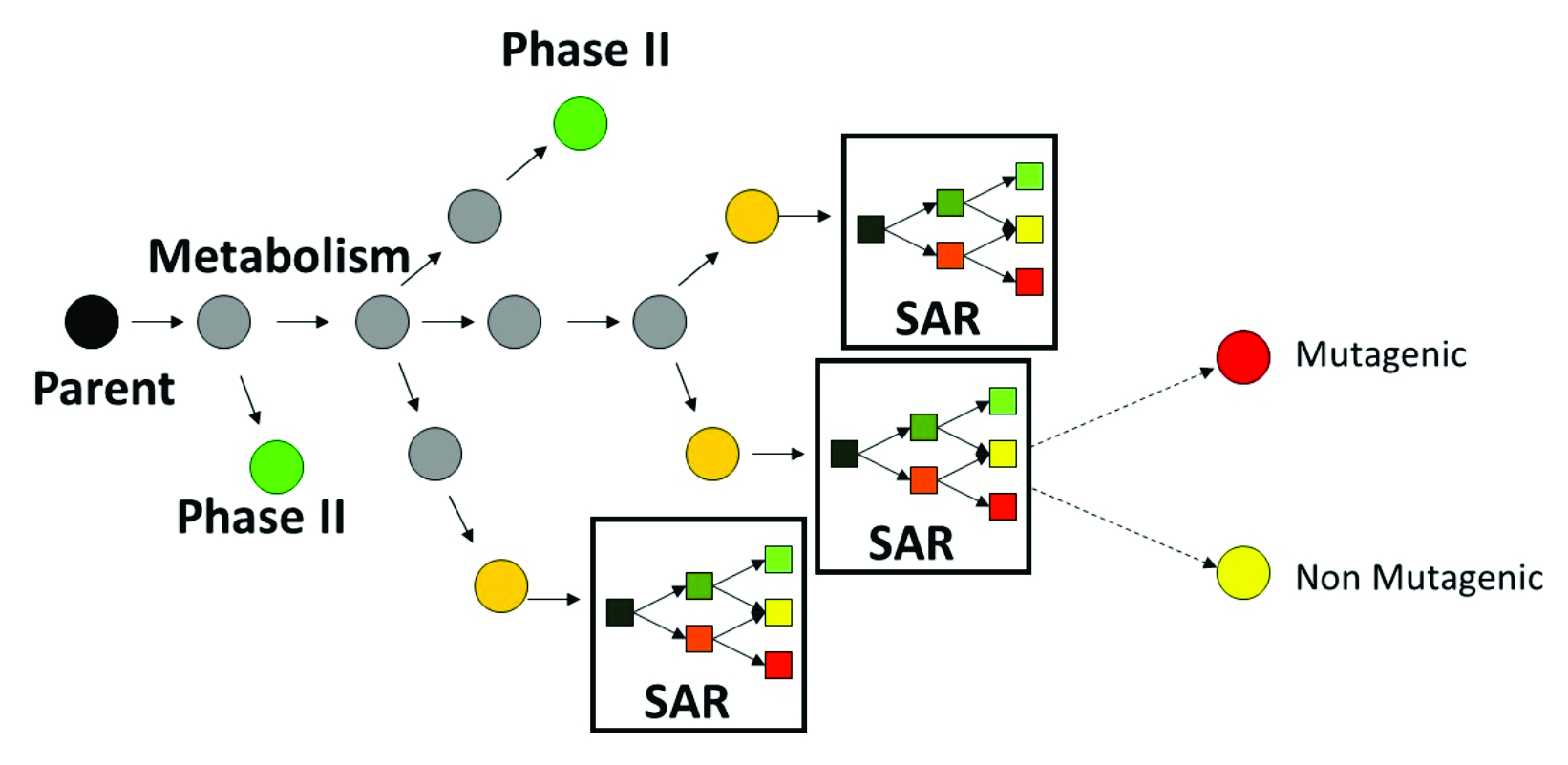
Figure 1. Illustration of the integral MLA-tk mutagenicity prediction based on metabolic simulator and defined structure-activity relationships (SARs).
Thus, chemicals are predicted to be mutagenic as parents only, parents and metabolites, and metabolites only.
According to the performance of TIMES_MLA-tk model (+S9) when applied on the training set chemicals, correct predictions are provided for:
- 180 predicted positives out of 197 observed positives after metabolic simulation give a sensitivity of = 91%,
- 93 predicted negatives out of 116 observed negatives after metabolic simulation give a specificity of 80%,
- 273 correct predictions out of the 313 observed positives and negatives after metabolic simulation give a concordance of 87%.
Domain
Each of the TIMES MLA-tk models (-S9 and +S9) has its own model applicability domain, which supports the MLA predictions [7]. The applicability domains consist of three sub-domain layers: general parametric requirements, structural features and alert(s) reliability. Two chemical subsets are used for deriving the model domains. The first subset includes the training set chemicals which are correctly predicted by the models, whereas the second subset comprises training set chemicals which are incorrectly predicted by the models.
The correct chemical subset is used for defining the general parametric requirements. Extracted are specific ranges of the molecular weight (MW) and the 1-octanol/water partition coefficient (log KOW):
• Molecular weight MW (in Da) ϵ [41; 1,256],
• log KOW ϵ [-10, 13].
The atom-centered fragments extracted from the correct subset of chemicals are used to define the structural domain. Briefly, the structural domain is assessed based on atom-centered fragments, extracted from correctly and incorrectly predicted (i.e., false positives and false negatives) substances from the model training sets by accounting for the atom type, hybridization and attached H-atoms of the central atom and its first neighbours. If the neighbour is a heteroatom then the diameter of the fragment is increased up to three consecutive heteroatoms or to the first carbon atoms in sp3 hybridization. In order to assess if a new chemical belongs to the structural domain, the system partitions the chemical to atom-centered fragments, which are then matched to the fragments extracted from the correct and incorrect chemical subsets. The new chemical is estimated to belong to the structural domain only when its atom-centered fragments are found in the list of correct fragments.
The third level of the domain accounts for reliability of alerts. Currently, information for alerts reliability is provided in the model reports.
Reporting
The predictions of in vitro MLA-tk model could be reported as:
- A tab delimited text file providing the following information for chemical: identity (CAS number, Name, SMILES), observed MLA-tk mutagenicity, predicted MLA mutagenicity, information for the alert(s) responsible for DNA and/or protein interaction, alert reliability and performance, applicability domain, etc.
- QSAR Prediction Reporting Format (QPRF) is available, generating a report for one chemical or group of chemicals.
The TIMES MLA-tk (+S9) model provides information for:
- Simulated metabolic map,
- Positive parents and/or metabolites,
- Mechanisms of DNA and/or protein damage,
- Local training set of alerts,
- Performance of alerts,
- Different types of reporting
- …..
Decision support character of TIMES_MLA-tk models
Users of the TIMES model may assess reliability of the predictions by evaluating the amount of mechanistic justification supporting the ultimate prediction. In this respect, the modeling results could be considered as decision supporting rather than decision making outcome.
References
- Applegate, M. L., Moore, M. M., Broder, C. B., Burrell, A., Juhn, G., Kasweck, K. L., Lin, P. F., Wadhams, A. and Hozier, J. C. (1990) Molecular dissection of mutations at the heterozygous thymidine kinase locus in mouse lymphoma cells. Proc. Natl Acad. Sci. USA, 87, pp. 51-55.
- Moore, M. M., Harrington-Brock, K., Doerr, C.L., Dearfield, K.L. (1989) Differential mutant quantitation at the mouse lymphoma tk and CHO hgprt loci. Mutagenesis, 4(5), pp. 394-403.
- Honma, M., Zhang, L.S., Sakamoto, H., Ozaki, M., Takeshita, K., Momose, M., Hayashi, M., Sofuni, T. (1999). The need for long-term treatment in the mouse lymphoma assay, Mutagenesis 14(1): 23-29.
- Myhr B.C., Caspary W.J. (1988) Evaluation of the L5178Y mouse lymphoma cell mutagenesis assay: intralaboratory results for sixty-three coded chemicals tested at Litton Bionetics, Inc. Environ. Mol. Mutagen. 12 (13): 103-194.
- Caspary, W.J., Daston, D.S., Myhr, B.C., Mitchell, A.D., Rudd, C.J., Lee, P.S. (1988) Evaluation of the L5178Y mouse lymphoma cell mutagenesis assay: Interlaboratory reproducibility and assessment, Environ. Mol. Mutagen 12: 195-229.
- Mekenyan, O., Dimitrov, S., Pavlov, T., Dimitrova, G., Todorov, M., Petkov, P., Kotov, S. 2012. Simulation of chemical metabolism for fate and hazard assessment. V. Mammalian hazard assessment, SAR and QSAR in Environmental Research, 23:5-6, 553-606.
- Dimitrov, S., Dimitrova, G., Pavlov, T., Dimitrova, N., Patlewicz, G., Niemela J., Mekenyan, O. 2005. J. Chem. Inf. Model., Vol. 45, pp. 839-849.
