Skin Metabolism
Endpoint
Skin metabolism simulator is part of TIMES Skin sensitization model and it mimics the metabolism of chemicals in the skin compartment.
Data
Given the lack of reported skin metabolism data and the widespread hypotheses that skin enzymes can metabolize absorbed xenobiotics via reactions analogous to those determined in liver, initially the simulator was developed as a simplified mammalian liver metabolism simulator. Currently, the simulator was upgraded and adjusted to simulate the documented in vitro metabolism of 151 chemicals.
Model
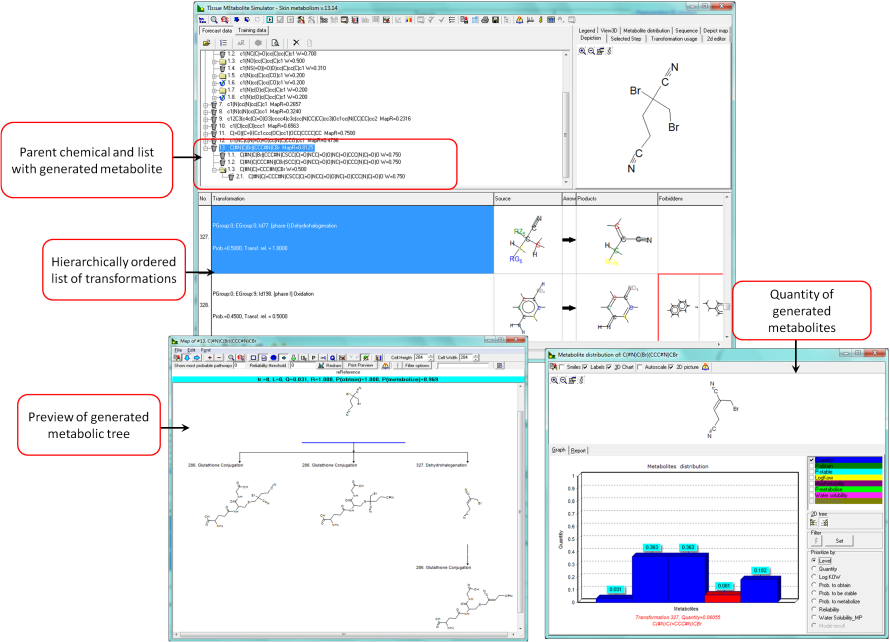
The skin metabolism simulator [1] contains a list of hierarchically ordered principal transformations, which can be divided into two main types - rate-determining and non-rate-determining. The rate-determining transformations are Phase I and Phase II, such as C-hydroxylation, ester hydrolysis, oxidation, glutathione conjugation, glucuronidation, sulfonation. The non-rate-determining transformations include hydrolysis of salts and molecular transformations of highly reactive intermediates. Each transformation in the simulator consists of source and product structural fragments, and inhibiting "masks". A probability of occurrence is ascribed to each transformation, which determines its hierarchy in the transformation list. Due to the limited qualitative and quantitative skin metabolism data reported for chemicals the reliabilities of transformations were assigned on the basis of available information or expert knowledge. The highest confidence level of 1.0 was assigned when the transformation was well established and supported by literature sources. A level of 0.75 was set when no reference source supporting the transformation existed, but the enzymatic reaction was nevertheless well known. A level of 0.5 was assigned when the transformation was expected, but the enzymatic system was unknown and a level of 0.25 when only expert judgment supported its feasibility.
Domain
The stepwise approach [2] was used to define the applicability domain of the model. It consists of the following sub-domain levels:
- General parametric requirements - includes ranges of variation log KOW and MW
- Structural domain - based on atom-centered fragments (ACFs)
Statistics
Average Sensitivity: S = [X/(X + Y)].100 [%]
Average Predictability: P = [X/(X + Z)].100 [%]
X: number of correctly generated (predicted) metabolites, observed also experimentally ("true positives");
Y: number of non-generated (non-predicted) but experimentally observed metabolites ("false negatives");
Z: number of generated but not experimentally observed metabolites ("false positives").
Simulator performance parameters:
Average sensitivity: S = 86.7 %
Average predictability: P = 34.7 %
Reporting
The model provides results for:
- Quantities of parent and metabolites, mol/mol parent,
Reliability of predicted metabolites
Refrences
- O. Mekenyan, S. Dimitrov, T. Pavlov, G. Dimitrova, M. Todorov, P. Petkov and S. Kotov. Simulation of chemical metabolism for fate and hazard assessment. V. Mammalian hazard assessment. SAR and QSAR in Environ Res, 23, (2012), pp. 553-606.
- S. Dimitrov, G. Dimitrova, T. Pavlov, N. Dimitrova, G. Patlevisz, J. Niemela and O. Mekenyan, J. Chem. Inf. Model. Vol. 45 (2005), pp. 839-849.