Criteria for quantitative assessment of metabolic similarity between chemicals. II. Application to human health endpoints
Computational Toxicology Volume 19 (2021) 100173
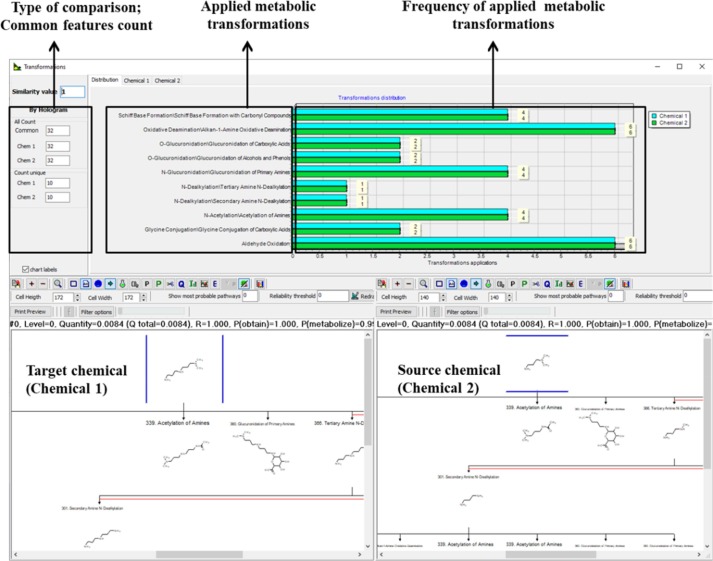
Identifying a suitable source analogue for a particular target chemical is the critical tasks in the development of a read-across prognostication. Although chemical pairings for read-across data gap filling can be selected by various similarity methods, it is increasingly important that the read-across documentation considered toxicokinetic parameters, especially metabolic ones in their similarity arguments. Case studies have confirmed that estimating similarity between metabolic pathways of a target and its analogue(s) is of high importance for corroborating a read-across prediction for human health effects. Previously, side-by-side comparisons of metabolic maps were demonstrated to be important in establishing metabolic similarity. Here are described automated modules that provide transparent information regarded key factors for confirming metabolic similarity between read-across analogues. Specifically, an in silico functionality dealing with several key aspects of metabolic similarity has been developed - tools to assess commonality 1) in prioritized metabolic transformations or reactions, in fingerprint or hologram type analysis of 2) patterns of reactivity and 3) structural patterns, as well as 4) in the availability of a toxic metabolite from dissimilar parent compounds. The advantages of evaluating the results from this new functionality are shown with four examples of human health effects. In each case, comparing the new metabolic similarity parameters between analogues identifies suitable read-across chemical pairings.