Assessing metabolic similarity for read-across predictions
Computational Toxicology Volume 18 (2021) 100160
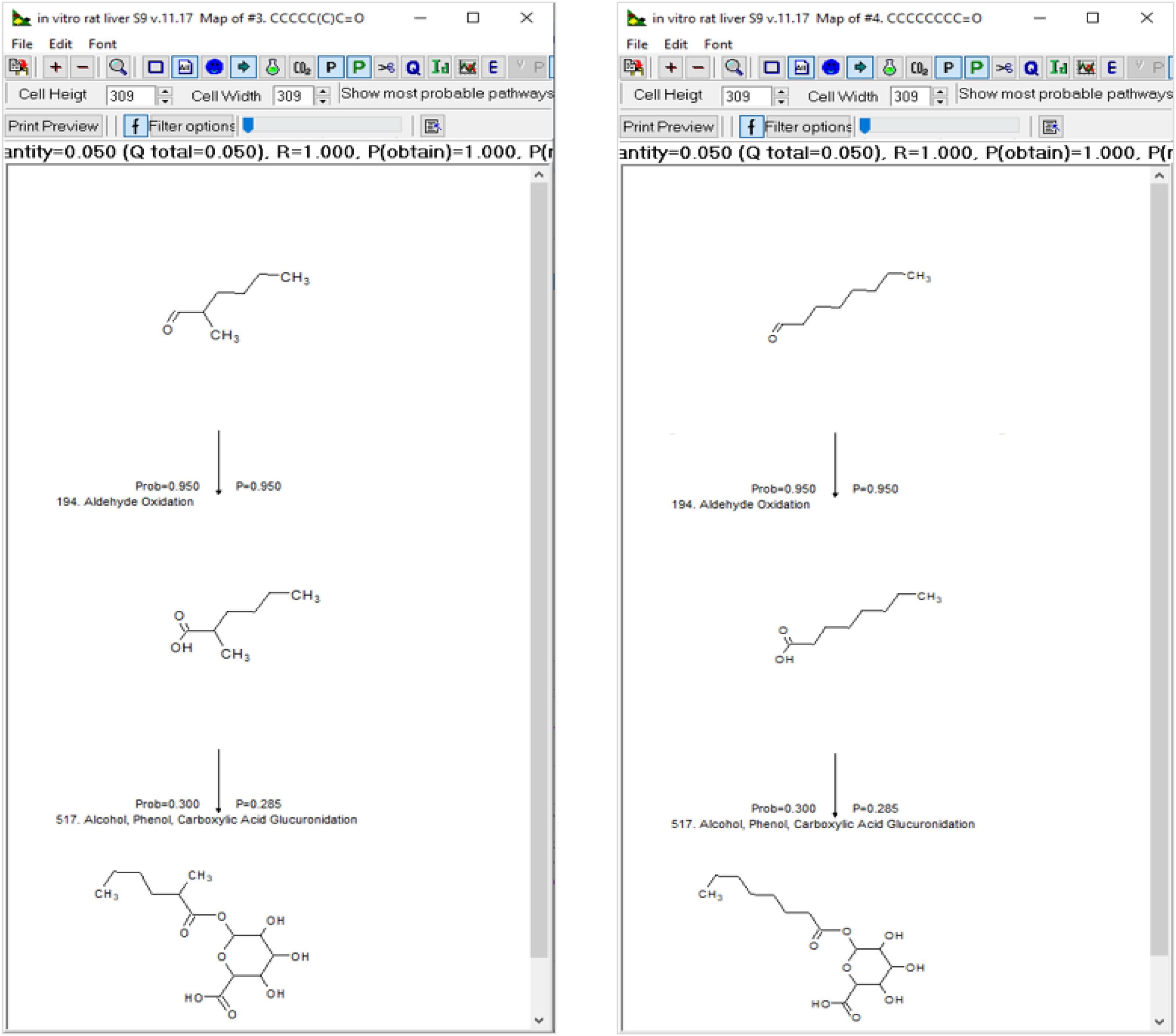
Read-across is one of the most frequently used non-animal approaches for filling toxicological data gaps. There is a universal realization that considerations of the metabolic fates of the target substance and proposed source analogue(s) are important in accepting or rejecting a read-across hypothesis. Comparisons of target and source metabolic information most often suffers from the lack of experimental data. These data voids have prompted the development and use of alternative methods. With various limitations, several in silico tools are available to aid an evaluator in appraising metabolic similarity. Metabolic maps, arrays of 2D molecular structures of (a)biotic transformation products, allow for side-by-side visual comparison of stepwise routes of conversion of the target and source chemicals. There are two varieties of metabolic maps - documented and simulated. A document metabolic map is one derived for in vivo or in vitro experimental data. A simulated one is a map derived from metabolic pathway predictions of possible or probable metabolites coupled with expert judgment. Here, through a series of examples, the advantages of using the metabolic information, provided by the OASIS software, are exemplified. This will aid in establishing metabolic consistency between the chemicals used in a read-across argument (i.e., establish similarity in their (a)biotic transformations). Of particular importance are found to be the identification of common metabolic pathway(s), common metabolite(s), and similarity concerning the formation of reactive metabolites.