Human Health Models
Q. Which Salmonella strains are included in building the Ames mutagenicity model?
A. Chemicals
included in the training set of the TIMES Ames mutagenicity model
are collected according to the recommendation in the OECD technical
guideline 471 addressing the number of Salmonella typhimurium
strains (TA100, TA98, TA1535, TA1537 (or TA97, TA 97A), E. coli WP2
uvrA (or E. coli WP2 uvrA (pKM101), or S. typhimurium TA102)
associated with each data:
- For negative effect, five Salmonella strains must show simultaneously negative data as described in the corresponding OECD guideline for testing of chemicals
- For positive effect, positive data in a single Salmonella strain would be enough
___________________________________________________________________
Q: What kind of results are provided by TIMES Ames mutagenicity model?
A: The model provides prediction results for the chemicals as parent structures and also predictions for all generated metabolites. The generated metabolic tree can be visually analizyed and in case of positive prediction supporting information of the interaction mechanism is provided.
___________________________________________________________________
Q: How can I speed up a file loading??
A: By default, when a file is loaded, all 2D and 3D properites of the structure in the file are calculated. Thus, the structures could be loaded quickly if their properties are not preliminary calculated. To do this, follow the steps described below:
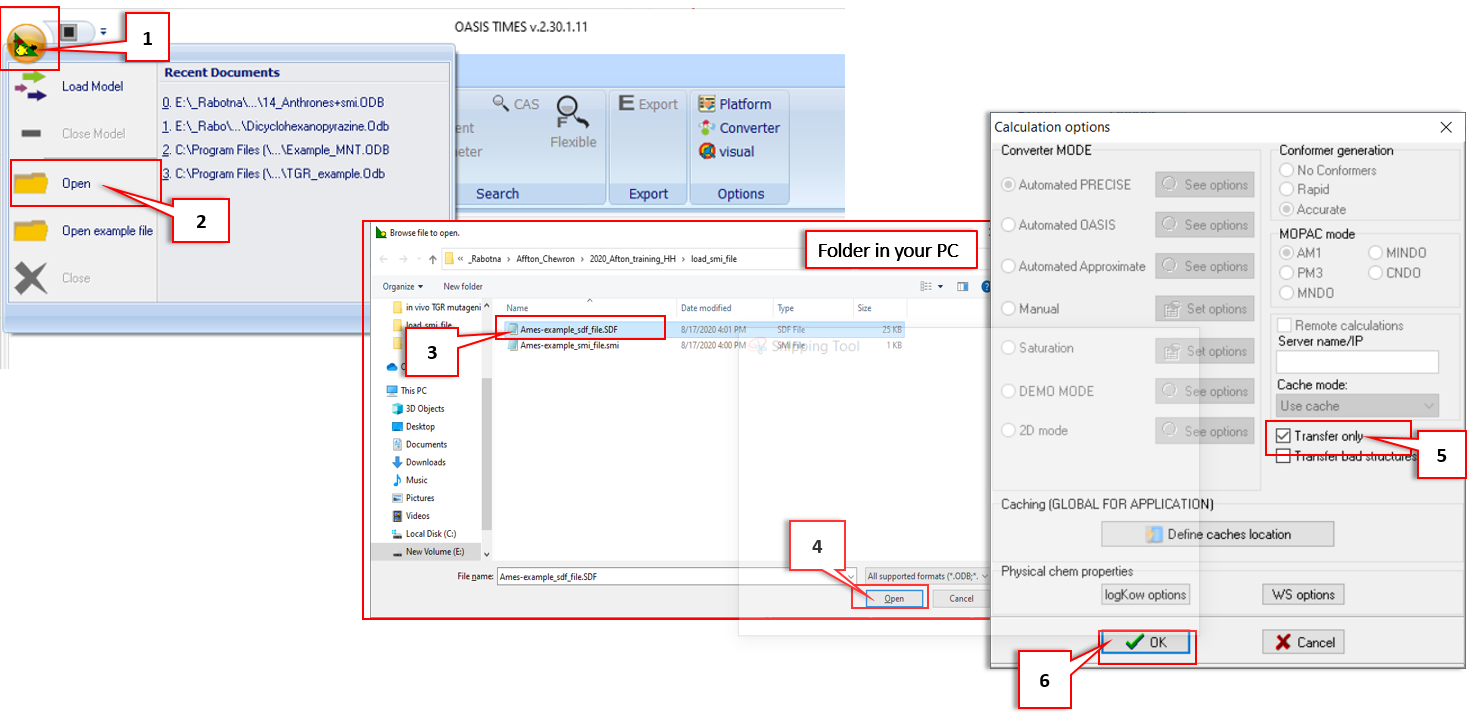
The important step here is #5. If the "Transfer only" option is checked, then the structures in the file will be loaded in TIMES automatically without any preliminary calculations.
__________________________________________________________________
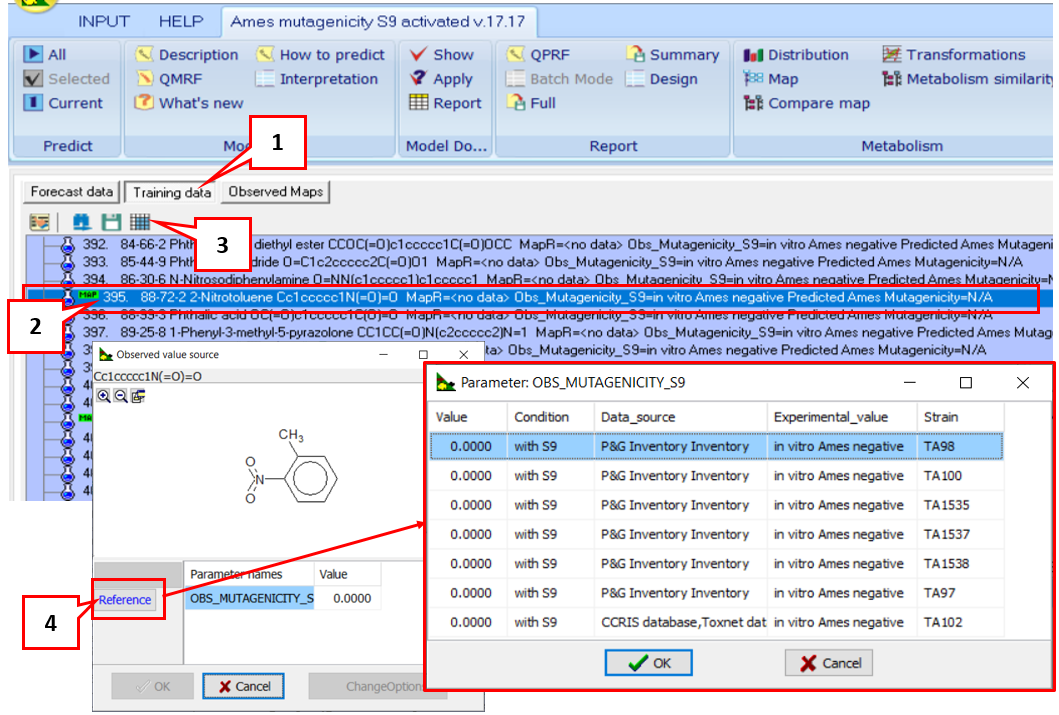
Q: How to see more details on the experimental Ames data?
A: If a chemical of interest has experimental data in TIMES, more details on it, could be seen by: 1) Going to the training set; 2) Click over the chemical; 3) Click on the icon for the observed value source; 4) Click Reference. A table with details on the experimental data - reference source, tested strain(s) will be shown.
Note: Acording to the TG 471, all chemicals with Negative Ames experimental data are confirmed with experimental Negative data for at least five different strains.
-------------------------------------------------------------------------------------------------------------------
Q: How to dock TIMES to OECD QSAR Toolbox?
A: In order to dock TIMES to Toolbox, you need just to install the latest official versions of both software (i.e. TIMES v.2.31.2 and QSAR Toolbox v.4.5).
Once, you have installed both on your computer, they will be automatically connected.
In this way, you will be able to 1) make a predictions within the Toolbox environment by the TIMES models and 2) to use the knowledge available in Toolbox (e.g. profilers) for searching chemicals with observed experimental data points or observed metabolic maps available in TIMES.
