Is the simulated metabolism adequate?
Let`s search for metabolism support!
Given the importance of the metabolism for the (Q)SAR predictions, the adequacy of the simulated metabolism should be evaluated, and the reliability of generated metabolites should be justified by documented metabolism data (if such data exist).
New functionalities have been developed allowing the users to support a simulated metabolic sequences leading to formation of specific metabolite.
This support could be:
- Experimental support - searching for analogues of the target chemical, having documented metabolic maps, where the same simulated metabolic sequence is reported;
- Theoretical support - searching for literature information justifying the simulated metabolic transformations
These functionalities been implemented in the OASIS TIMES and CATALOGIC software. The results are reported in the QSAR Prediction Reporting Format (QPRF).
Example:
Target chemical: 4-(tert-Butyl)phenyl isocyanate
Target endpoint: Bioaccumulation
Isocyanate hydrolysis is simulated on the first metabolism level for the target chemical by CATALOGIC BCF base-line model.
As the first metabolism level is crucial for the bioaccumulation, we may want to support this transformation.
- Experimental support
To support the simulated transformation sequence (isocyanate hydrolysis) the documented analogues should:
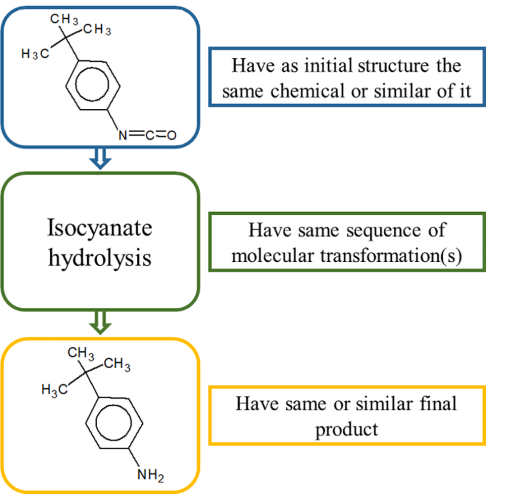
The analogues are searched in the database with chemicals having documented metabolic maps, available in the CATALOGIC software.
One analogue meeting the defined criteria is found in the CATALOGIC`s database:
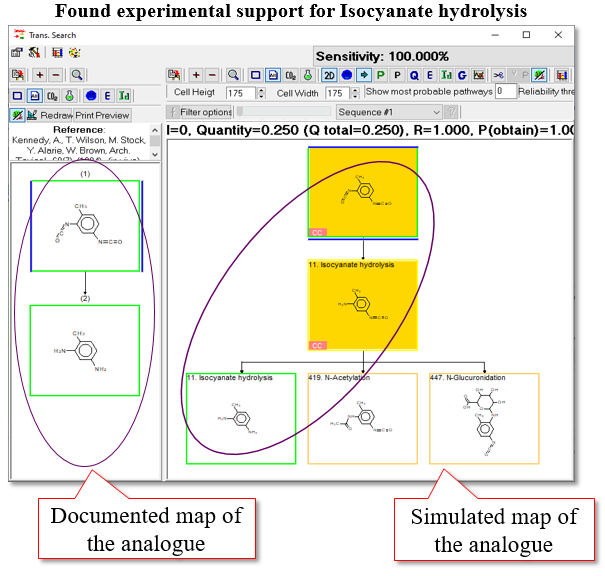
- Theoretical support
Library with literature information for the metabolic transformations has been preliminary collected and included in the CATALOGIC software.
Information for hydrolysis of isocyanates is also availble in the library:
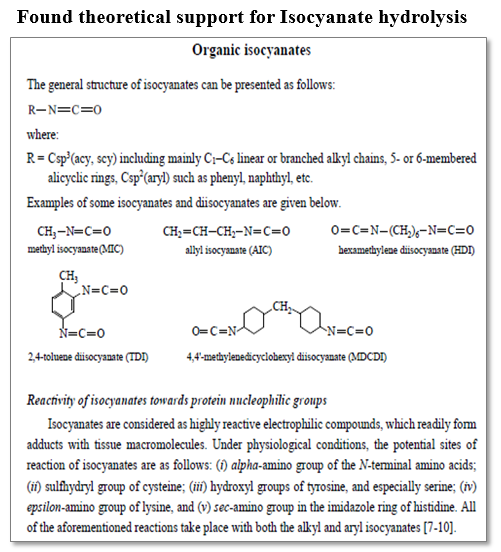
Similar analysis and support could be done for each of the simulated metabolites/metabolic sequences by the OASIS (TIMES and CATALOGIC) models.
