CATALOGIC 5.15.2 released
February, 2022
CATALOGIC 5.15.2 (Feb, 2022) is available for download.
I. Modifications in the platform
- The CATALOGIC software requires activation of a License key with an expiration period (depending on the contractual issues) to allow working with the system.
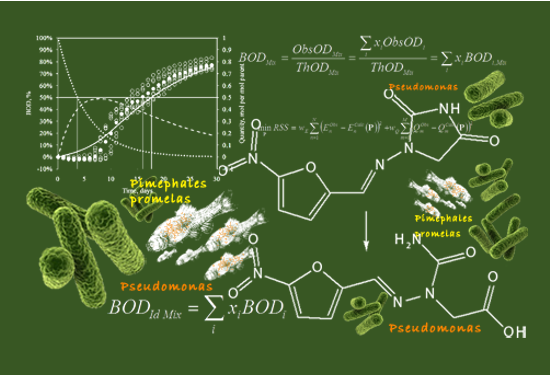
- Documented Metabolism database version is updated from 3.1.1 to 3.4.0 (Application (MetaPath) version 5.4.0).
- The list with cache files used for storing of pre-calculated results has been extended to accelerate the work with the system.
- EPIWin v.4.11 module is used as default calculations of logKow, Molecular weight and Water solubility parameters accounted in the parametric layer of the applicability domain.
- In case the calculation from EPIWin v.4.11 cannot be done, the old dll module (Syracuse University) is applied.
- The panel displaying the current chemical detailed information is significant redesigned and extended with context specific actions.
- Docking with OECD QSAR Toolbox version 4.5
II.
Modifications in the models, existing and new
functionalities
- The prediction workflow has been improved organizing all the functionalities associated with explain of the predictions, searching for analogues supporting prediction, analogues supporting simulated metabolism, domain information and reporting.
- New functionalities have been introduced in the software for:
o Evaluating adequacy of
simulated metabolism by providing:
-Experimental
support - finding analogues with documented
metabolism
data based on selected criteria supporting selected sequence of
simulated
transformations
-Theoretical
support - mechanistic justification of the
transformations
simulating metabolism (help files have been developed for
biodegradation and bioaccumulation models)
o Clustering -
grouping of chemicals based on selected criteria
o Similarity matrix - contingency table for
the similarity between chemicals based
on selected criteria
o Selection of representatives -
prioritization of the chemicals based on their
metabolic/structural similarity compared to the other chemicals in
the list
o Entropy - removing less informative
chemicals based on selected criteria
o Metabolism
similarity - calculate similarity between chemicals
accounting
different metabolic features
o Option to predict metabolites, which pass
dedicated filter.
- Help files with more detailed information are provided within the software platform.
- Flexible search functionality has been extended with:
o Search by list (SMILES, Chemical name)
o Search by Distance for Parameters
o Masks could be added if fragment search is
used
o Transformation
search
- Improvements in the QPRF of the models - new organization of generation window, new appendix for analogues supporting metabolism, spell-check included for userdefined sections, save/load functionalities implemented.
- The categorization of transformations` names has been improved for BCF base-line models.
- Domain is not applied on hydrolysis products in models accounting for metabolism.
III. New
models
• Acidic hydrolysis model at pH 5
CATALOGIC
environmental fate and ecotoxicity