CATALOGIC 5.17.1 released
July, 2024
CATALOGIC 5.17.1 (July, 2024) is available for download.
I. Modifications in the platform
-
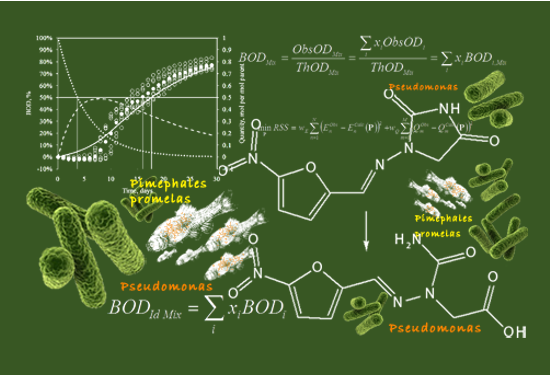
Documented Metabolism database version is updated from 3.4.1 to 3.4.2 (compatible with MetaPath version 5.4.2).
- The speed of calculation of EPI Win logKow and Water solubility parameters has been significantly accelerated.
- The Load toxicity models functionality has been moved to the INPUT tab.
- Ability to load multiple toxicity models at once.
- The Report functionalities have been moved to the FATE tab.
- The new option “Adaptive Probability to Obtain” threshold enables dynamic change of the probability threshold, confining the propagation of the metabolic map. This allows metabolization to be simulated adequately for large symmetric molecules.
- The new option “Merge equal branches” removes the duplicated equal branches. Instead of all, only one branch remains, representing the duplicated branches. The count of merged branches is the property of the representative branch. The number is also used in the metabolism formalism to correctly calculate the properties (e.g., quantities) of resulting metabolites in the same way as when all the branches are presented separately. So, this new option does not affect the calculations of metabolic properties but improves the 2D visualization of the simulated metabolic map and uses less memory.
- New applicability domains have been extracted for all models because of the new version of Domain Manager.
- Docking with OECD QSAR Toolbox v.4.7.
II. Modifications in the models, existing and new functionalities
- Modifications in biodegradation models:
o The performance of the CATALOGIC 301C model was improved with respect to simulated metabolism and predicted BODs of monomers and iso-olefins.
- Modifications in bioaccumulation models:
- Modifications in hydrolysis models:
o The training set of the Hydrolysis pH5 model was expanded with 12 chemicals.
-
Improvements in the QPRF associated with QAF documentation – the updates are in accordance with the new QPRF template, taking into account all new and modified sections of the QAF documentation.
- Improvements in the QMRF associated with QAF documentation – the updates are in accordance with the new QMRF template, taking into account all new and modified sections of the QAF documentation.
- New functionality in the BCF models allows to change and save the bioaccumulation assessment threshold that appears in the BCF chart (FATE tab > Options > View options).
- Generation of customized QPRF is also allowed for the metabolic simulators where the tissue specific metabolism is considered only, with no relation to the endpoints.
- The visualization of transformations names is improved, i.e., the full transformation names are already visible on the simulated 2D map and reported in the QPRFs.
- Color legend is implemented in the 2D map with simulated metabolism to support the explanation of different highlighting.
- The Clustering functionality is expanded with new criteria for grouping of chemicals, based on 2D/3D parameters and custom structural fragment.
- The complex Flexible search combined with metabolism is already allowed to be explained (so far, flexible search without metabolism was allowed for explain only).
- The simulated metabolic maps could be already filtered based on different criteria and applying the Flexible search functionality (e.g., using the knowledge from Toolbox, 2D/3D parameters, structure similarity, etc.).
- New functionality showing the quantity distribution of parent/metabolites over the time is implemented in the Metabolite distribution window.
CATALOGIC
environmental fate and ecotoxicity