CATALOGIC Abiotic 301C
Endpoint
The biodegradation CATALOGIC Abiotic 301C model simulates abiotic aerobic biodegradation under MITI I (OECD 301C) test conditions. The modeled endpoint is the quantities (mol/mol parent) of the parent chemical and its transformation products at 28 days, as a result of abiotic transformations.
Data
The training set of the model consists of 132 chemicals and 252 experimental quantities for parent chemicals and their transformation products. The collected data were stored in the Laboratory of Mathematical Chemistry (LMC) file format for computer treatment of degradation pathways. Because the sequence of products formation was missing in the experimental data, the structure of the observed pathways was simplified to two level trees containing only a root corresponding to the parent and leaves corresponding to the products.
Model
A set of 128 abiotic molecular transformations was extracted from the experimental data. Hydrolysis of different functional groups such as carboxylic acid anhydrides, carbamates, phosphoric acid esters, iso(thio)cyanates, nitriles, and oxiranes dominate the molecular transformations. Oxidation, decarboxilation, dehalogenation and tautomeric reactions are also included in the model. Probabilities of 26 transformations were ascribed on the bases of expert knowledge since sufficient experimental data for their estimation were missing. The probabilities of the remaining transformations were were estimated by the non-linear least square method:
![]()
where QObs and QCalc are observed and predicted quantities, respectively, and P is a vector of estimated probabilities of transformations. Further details on the mathematical formalism of the model can be reviewed in [1, 2].
Domain
The stepwise approach [3] was used to define the applicability domain of the model. It consists of the following sub-domain levels:
- General parametric requirements - includes ranges of variation log KOW and MW,
- Structural domain - based on atom-centered fragments (ACFs).
A chemical is considered In Domain if its log KOW and MW are within the specified ranges and if its ACFs are presented in the training chemicals. The information implemented in the applicability domain is extracted from the correctly predicted training chemicals used to build the model and in this respect, the applicability domain determines practically the interpolation space of the model.
Performance
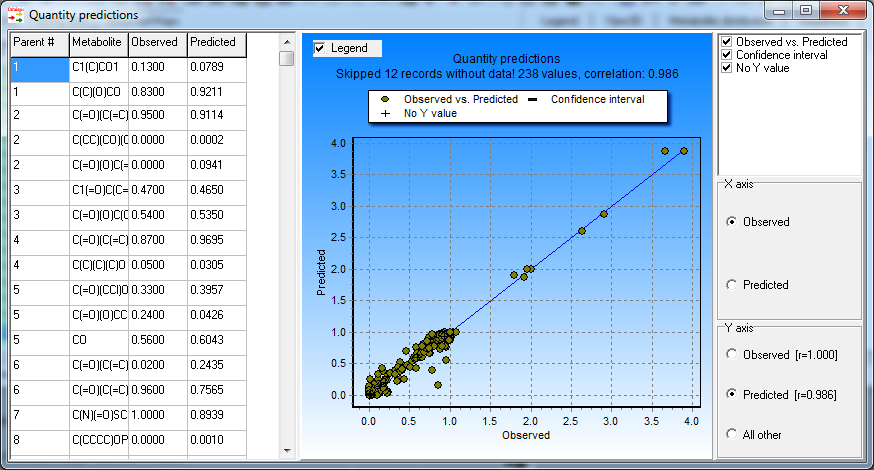
The goodness of fit evaluated by the squared coefficient of correlation is R2 = 0.97. No significant outlines are observed.
Reporting
The model provides results for:
- Primary half-life, days,
- Ultimate half-life, days,
- Quantities of parent and biodegradation products as a result of abiotic transformations, mol/mol parent,
Applicability domain details.
References
- S Dimitrov, T Pavlov, G Veith, O Mekenyan. SAR and QSAR in Environ Res, 22, 2011, 699-718.
- S Dimitrov, T Pavlov, N Dimitrova, D Georgieva, D Nedelcheva, A Kesova, R Vasilev, O Mekenyan. SAR and QSAR in Environ Res, 22, 2011, 719-755.
- S. Dimitrov, G. Dimitrova, T. Pavlov, D. Dimitrova, G. Patlevisz, J. Niemela and O. Mekemyan, J. Chem. Inf. Model. 45 (2005), pp. 839-849.
CATALOGIC Abiotic
Model features
Click the images for a larger view
Observed maps
Observed and predicted quantities
of training set chemicals
Model description pdf