CATALOGIC Kinetic 301F
Endpoint
The biodegradation CATALOGIC 301F model simulates aerobic biodegradation under OECD 301F test conditions. The modeled endpoint is the percentage of theoretical biological oxygen demand on 28th day (BOD, %).
Data
The training set contains 27860 kinetic BOD data for 979 proprietary chemicals under OECD 301F test conditions. The training set includes 562 readily biodegradable and 417 not readily biodegradable chemicals. Experimental half-lives were determined from the kinetic curves for 641 chemicals and were also used to parameterize the model.
Another training database of catabolic pathways for more than 551 organic compounds and expert knowledge was used to determine the principal transformations and to train the system to simulate aerobic catabolism of training chemicals. The documented pathways of microbial catabolism were collected from scientific papers, monographs and databases accessible over the Internet.
Model
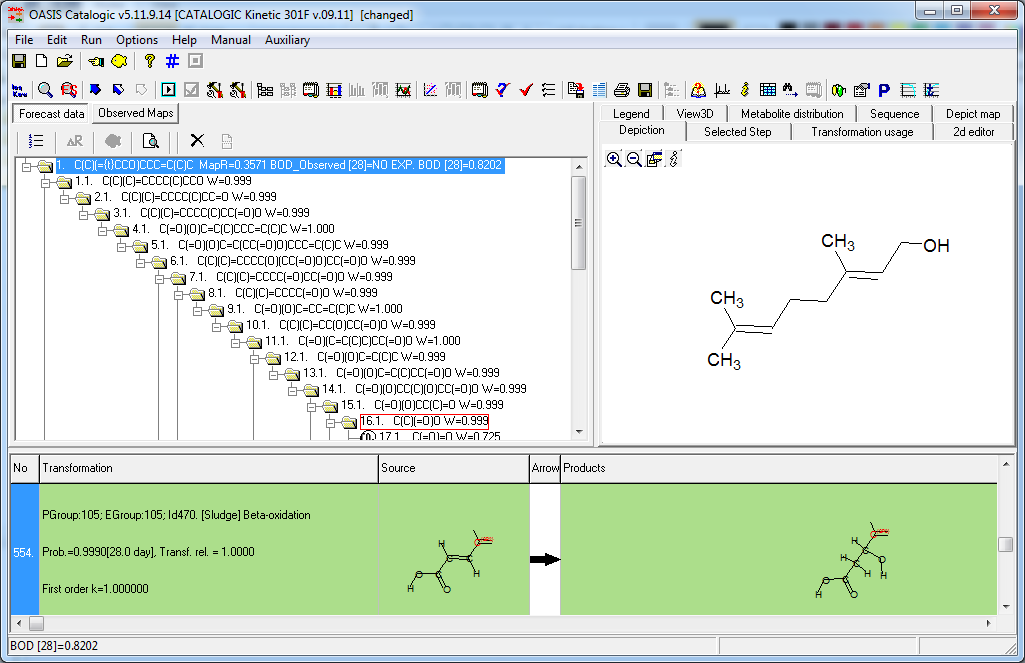
CATALOGIC 301F model consist of a metabolism simulator and an endpoint model. The microbial metabolism is simulated by the rule-based approach. The core parts of the simulator are a set of hierarchically organized transformations and a system of rules that control the application of these transformations. Recursive application of the transformations allows simulation of metabolism and generation of biodegradation pathways. Calculation of the modeled endpoint (BOD, %) is based on the simulated catabolic tree and the material balance of transformations used to build the tree.
The development of the model consists of: (i) generation of metabolic maps for the training set chemicals using the microbial metabolism simulator; (ii) estimation of probabilities of occurrence of the simulator transformations.
The probabilities were estimated assuming a first order kinetics:
![]()
where ki is a surrogate of the first order kinetic constant of the ith transformation. Non-linear least square fitting was used to parametrize the model:
![]()
where BODObs and BODCalc are observed and predicted BOD values, t1/2 is the ultimate half-life for nth chemical from the training set and k is a vector of estimated kinetic constant of the transformations. Further details on the mathematical formalism of the model can be reviewed in [1, 2].
Domain
The stepwise approach [3] was used to define the applicability domain of the model. It consists of the following sub-domain levels:
- General parametric requirements - includes ranges of variation log KOW and MW,
- Structural domain - based on atom-centered fragments (ACFs),
- Domain of simulator of metabolism - determines the reliability of the simulated metabolism.
A chemical is considered In Domain if its log KOW and MW are within the specified ranges, its ACFs are presented in the training chemicals and if the simulator contains transformations for its full mineralization. The information implemented in the applicability domain is extracted from the correctly predicted training chemicals used to build the model and in this respect the applicability domain determines practically the interpolation space of the model.
Performance
The goodness of fit evaluated by the squared coefficient of correlation and adjusted Pearson's contingency coefficient is R2 = 0.84 and C* =0.84, respectively. The model correctly classified 96% of experimentally ready and 81% of not ready degradable training chemicals.
Reporting
The model provides results for:
- BOD, %,
- Primary half-life, days,
- Ultimate half-life, days,
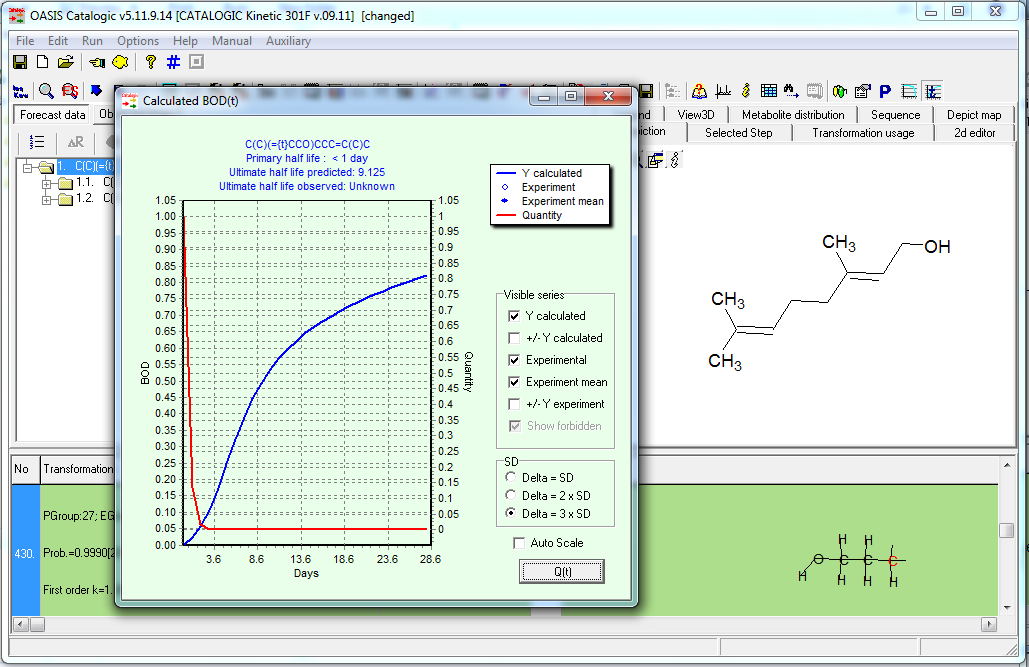
- Quantities of parent and biodegradation products, mol/mol parent,
- BOD, % within the 10 days window,
Applicability domain details.
References
1. S Dimitrov, T Pavlov, G Veith, O Mekenyan. SAR and QSAR
in Environ Res, 22, 2011, 699-718.
2. S Dimitrov, T Pavlov, N Dimitrova, D Georgieva, D Nedelcheva, A
Kesova, R Vasilev, O Mekenyan. SAR and QSAR in Environ Res, 22,
2011, 719-755.
3. S. Dimitrov, G. Dimitrova, T. Pavlov, N. Dimitrova, G.
Patlevisz, J. Niemela and O. Mekenyan, J. Chem. Inf. Model
CATALOGIC 301F
Model features
Click the images for a larger view
BOD, %
Biodegratation curve and quantity of parent
Model description pdf